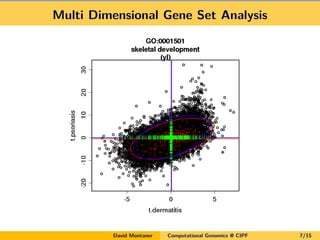
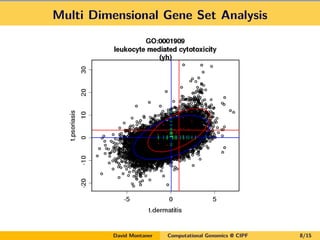
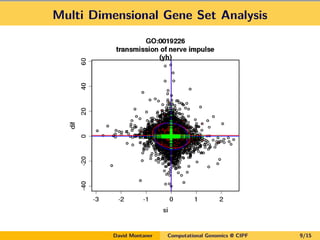
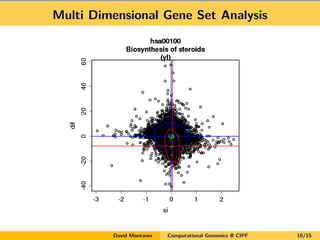
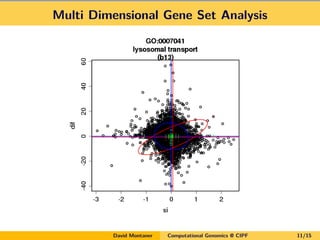
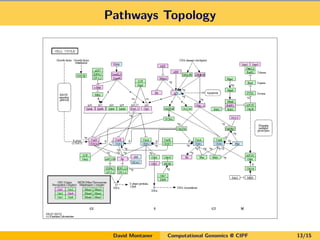
David Montaner is a statistician who works in computational genomics, focusing on massive data analysis and gene set analysis. He has developed methods for multi-dimensional gene set analysis and improving gene set analysis for next generation sequencing data. His ongoing work includes improving software implementation, adjusting methods for NGS data, extending the approach to other genomic features, and investigating topological pathway analysis and metagenomics.