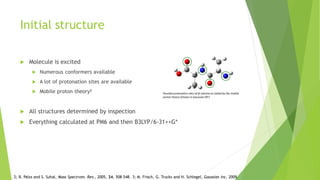
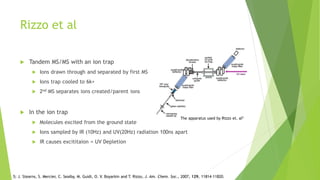
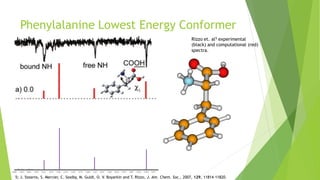
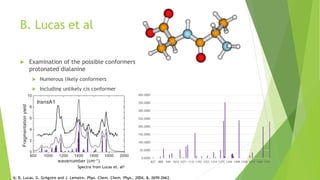
This document discusses computing infrared spectra of protonated peptides and fragments to compare with experimental data for identification. It summarizes previous work using tandem mass spectrometry and ion traps to obtain experimental IR spectra of peptides, as well as computational methods using PM6 and DFT to calculate IR spectra for comparison. Specific examples are provided of calculating spectra for phenylalanine and dialanine and comparing to experimental data.