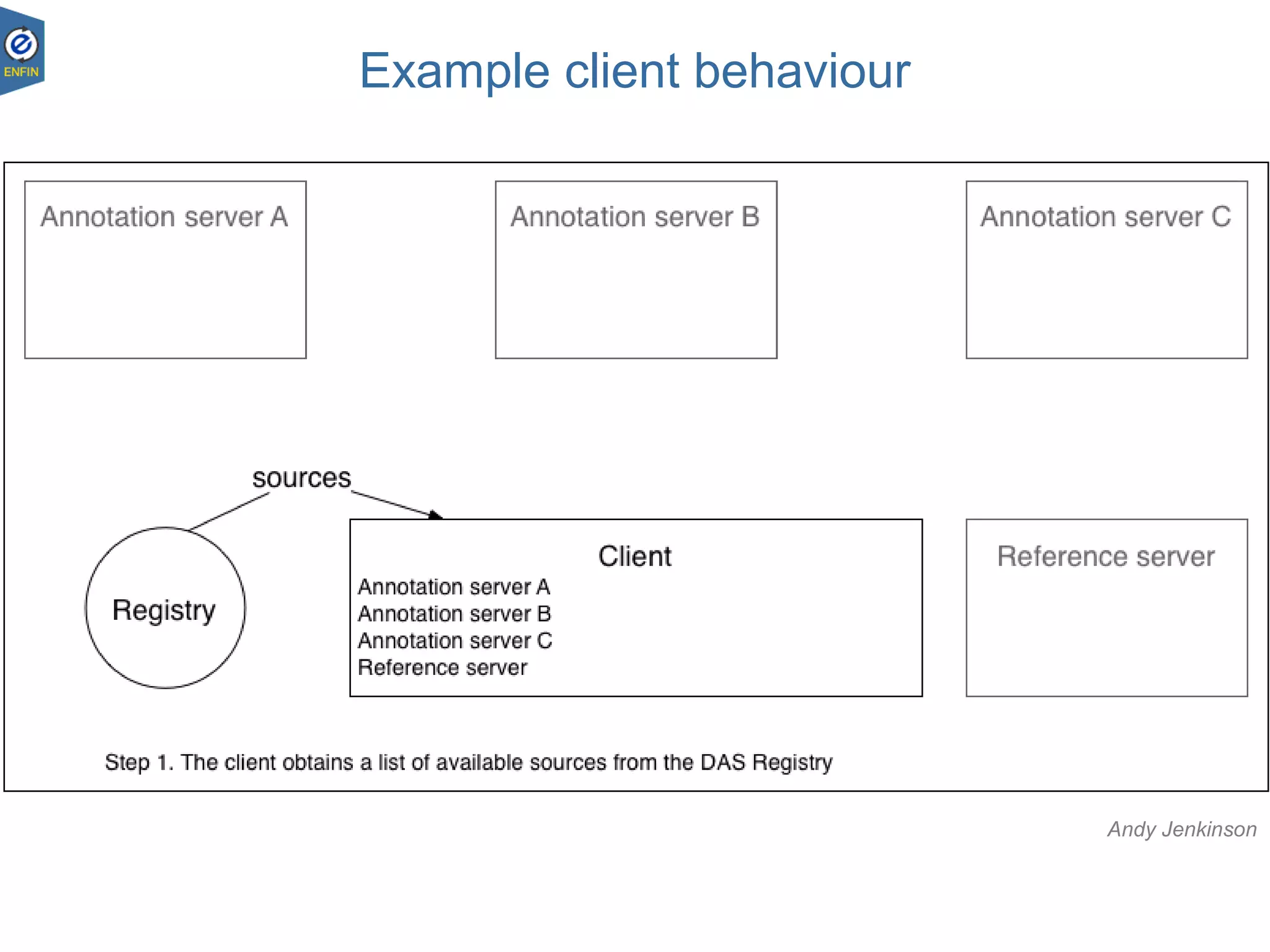
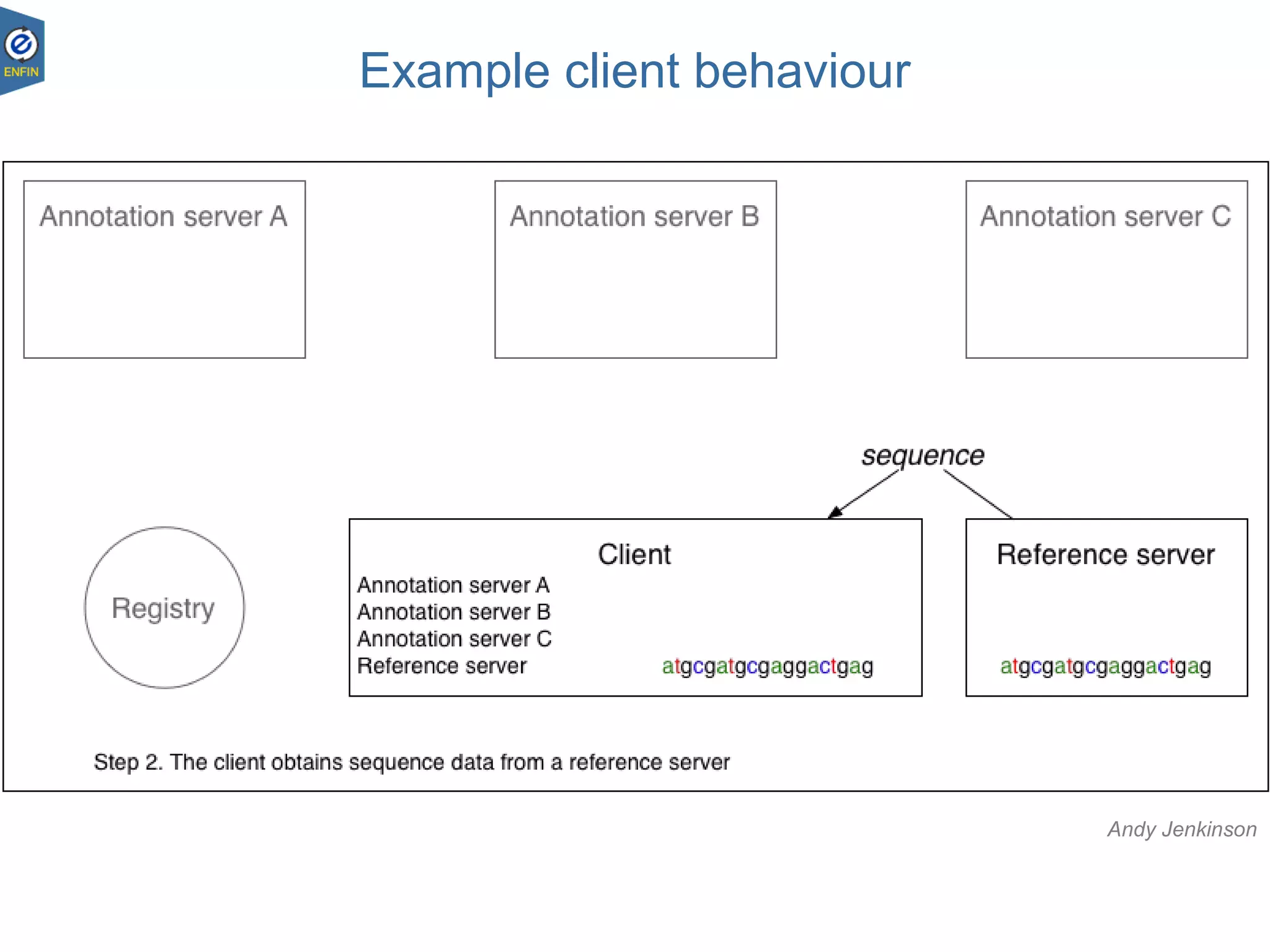
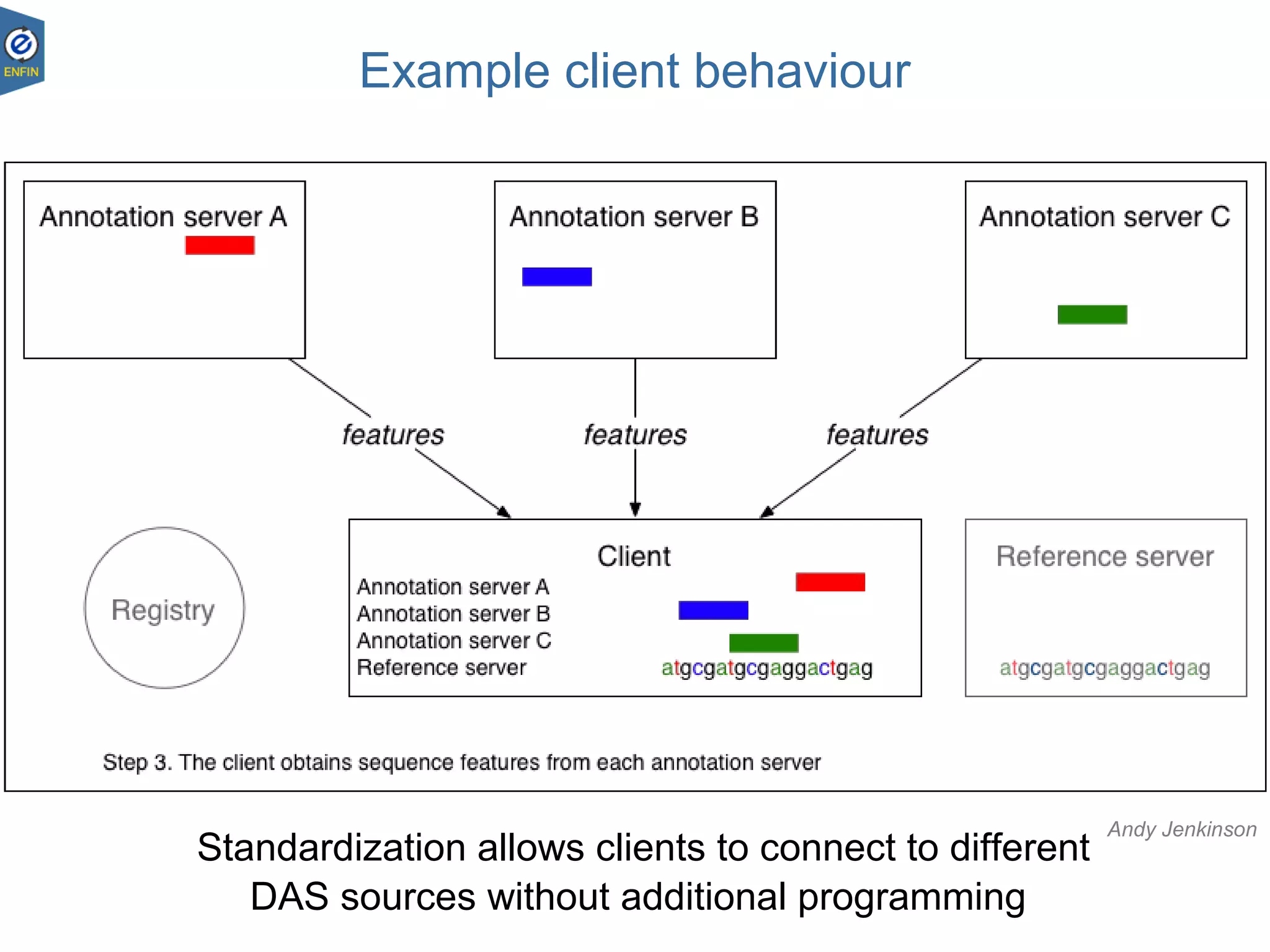
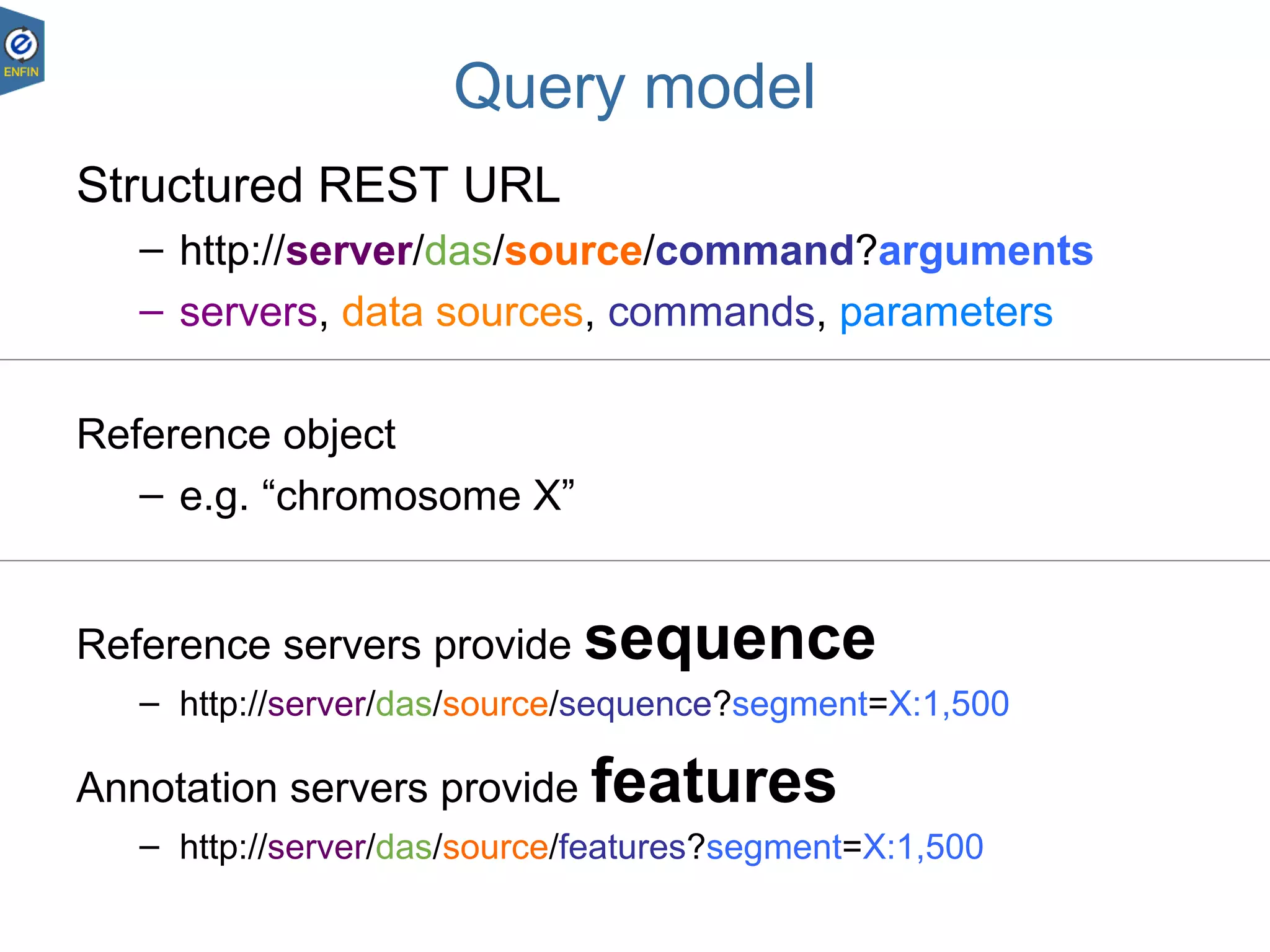
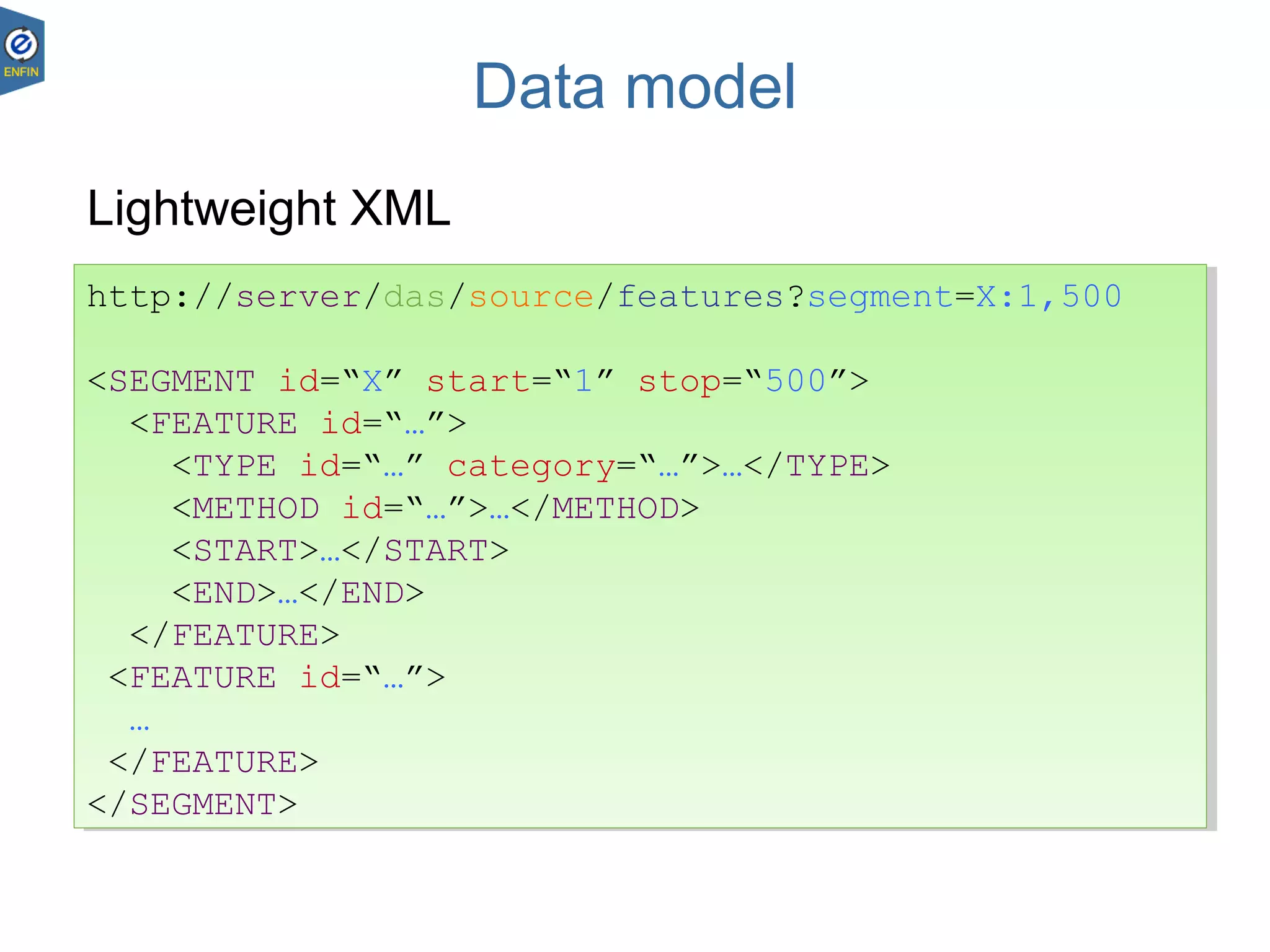
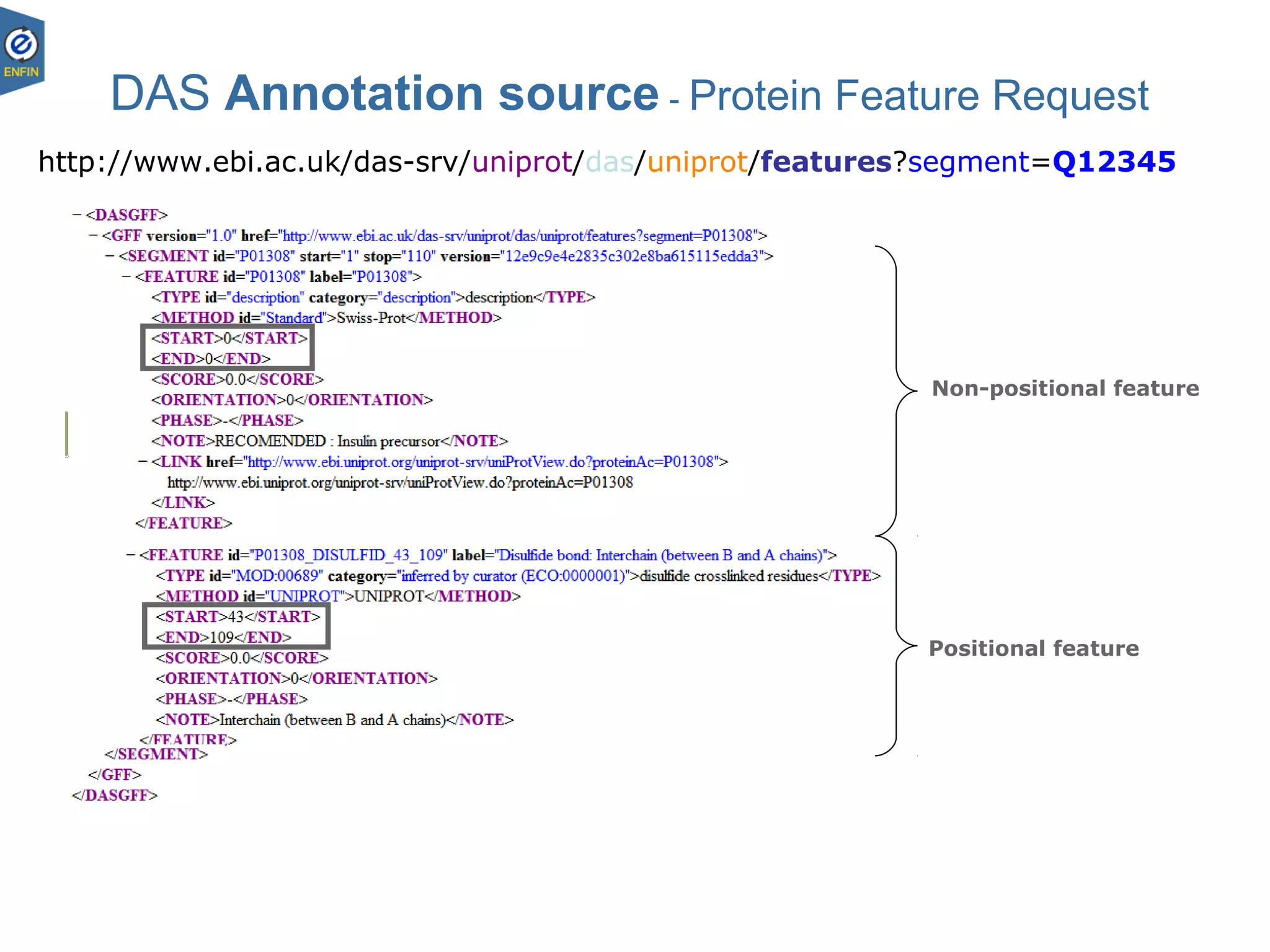
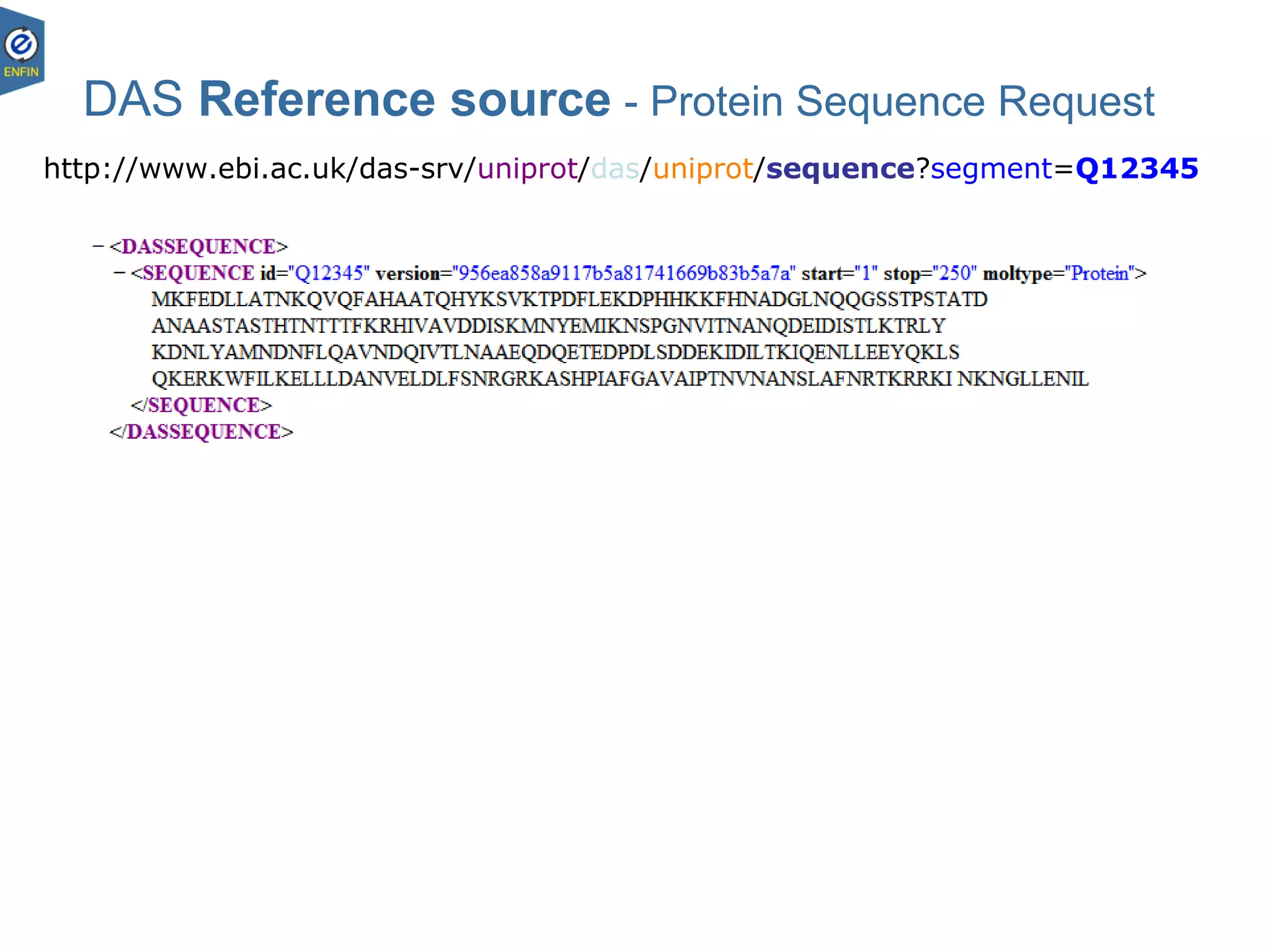
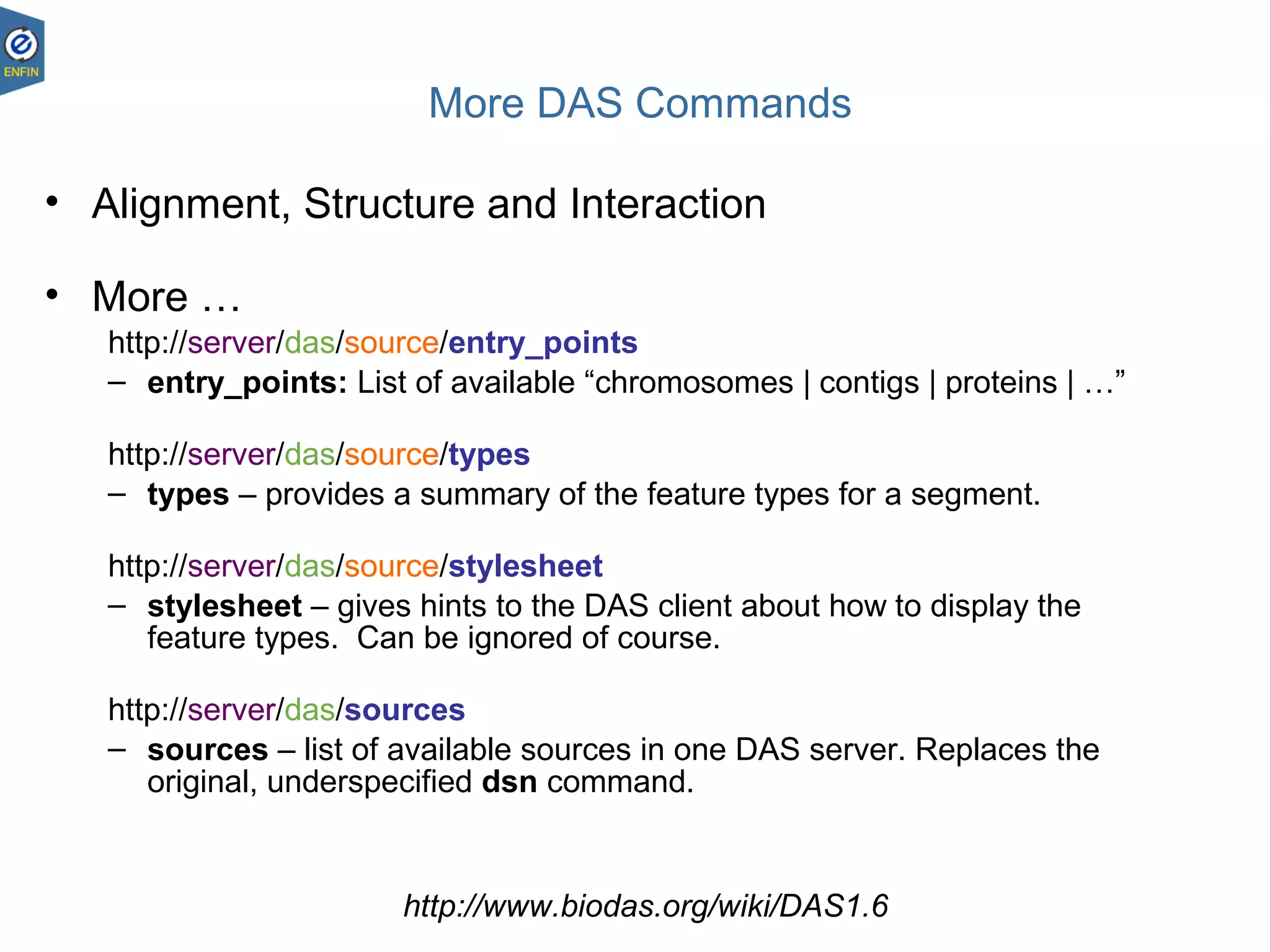
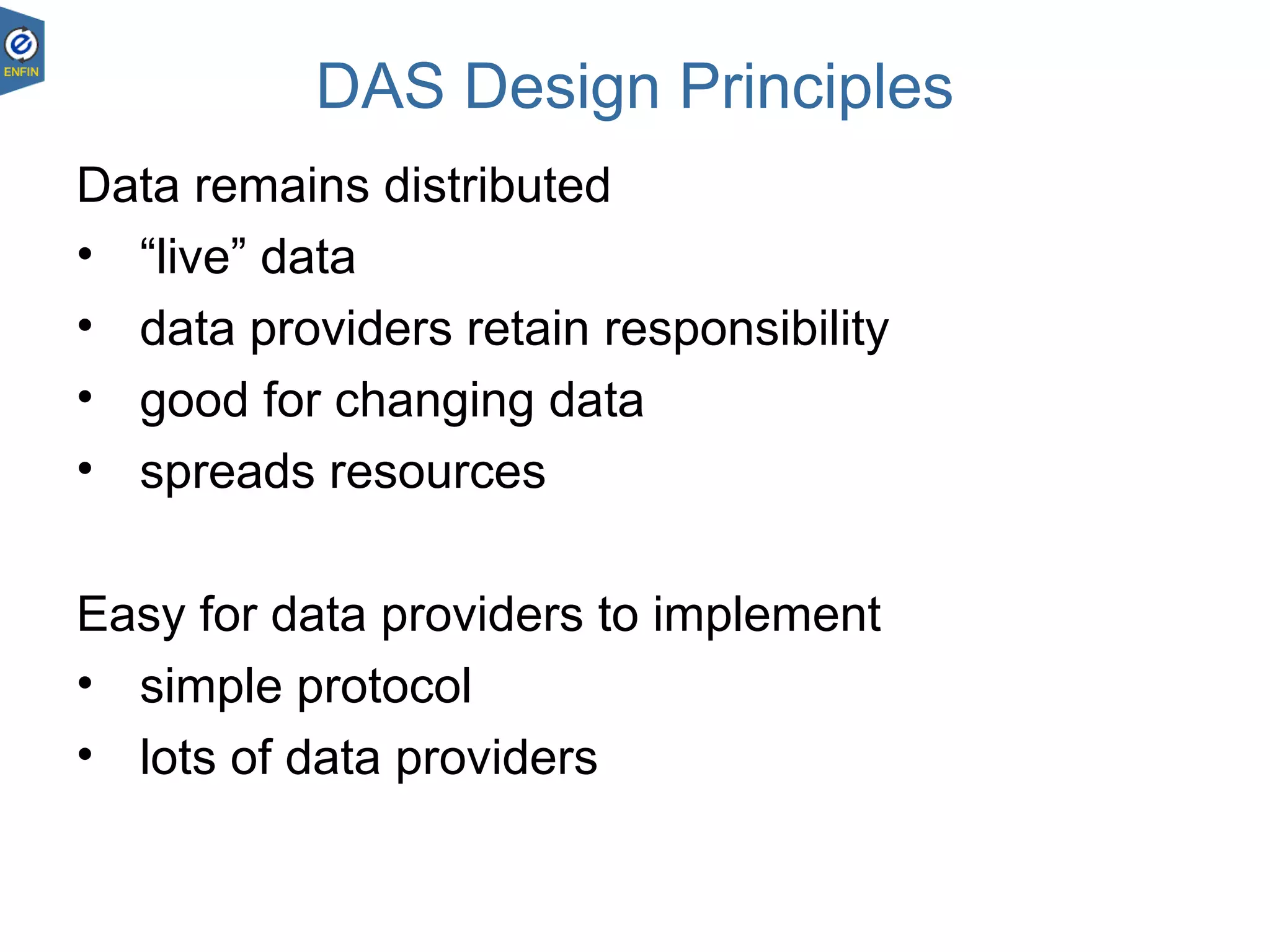
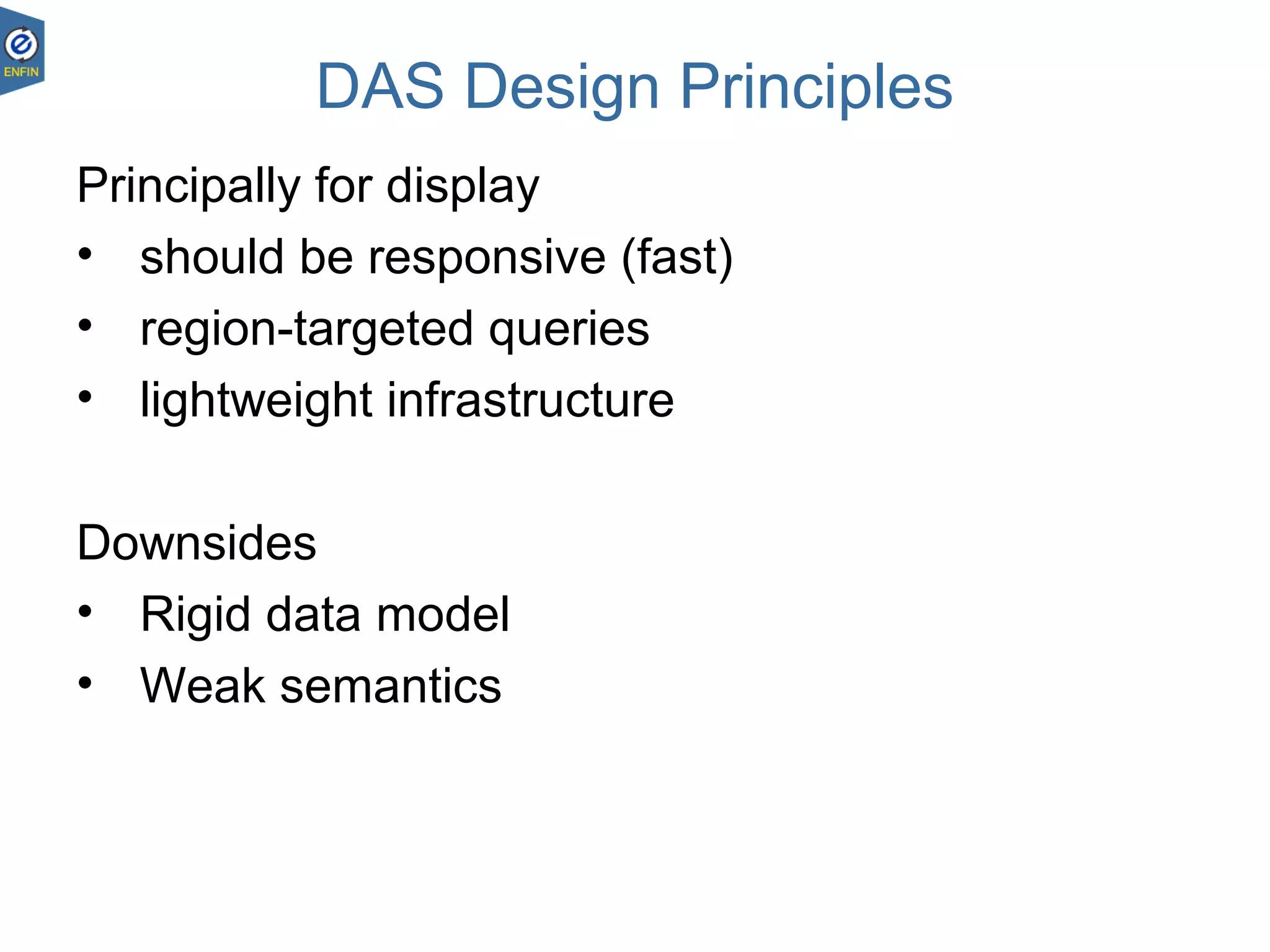
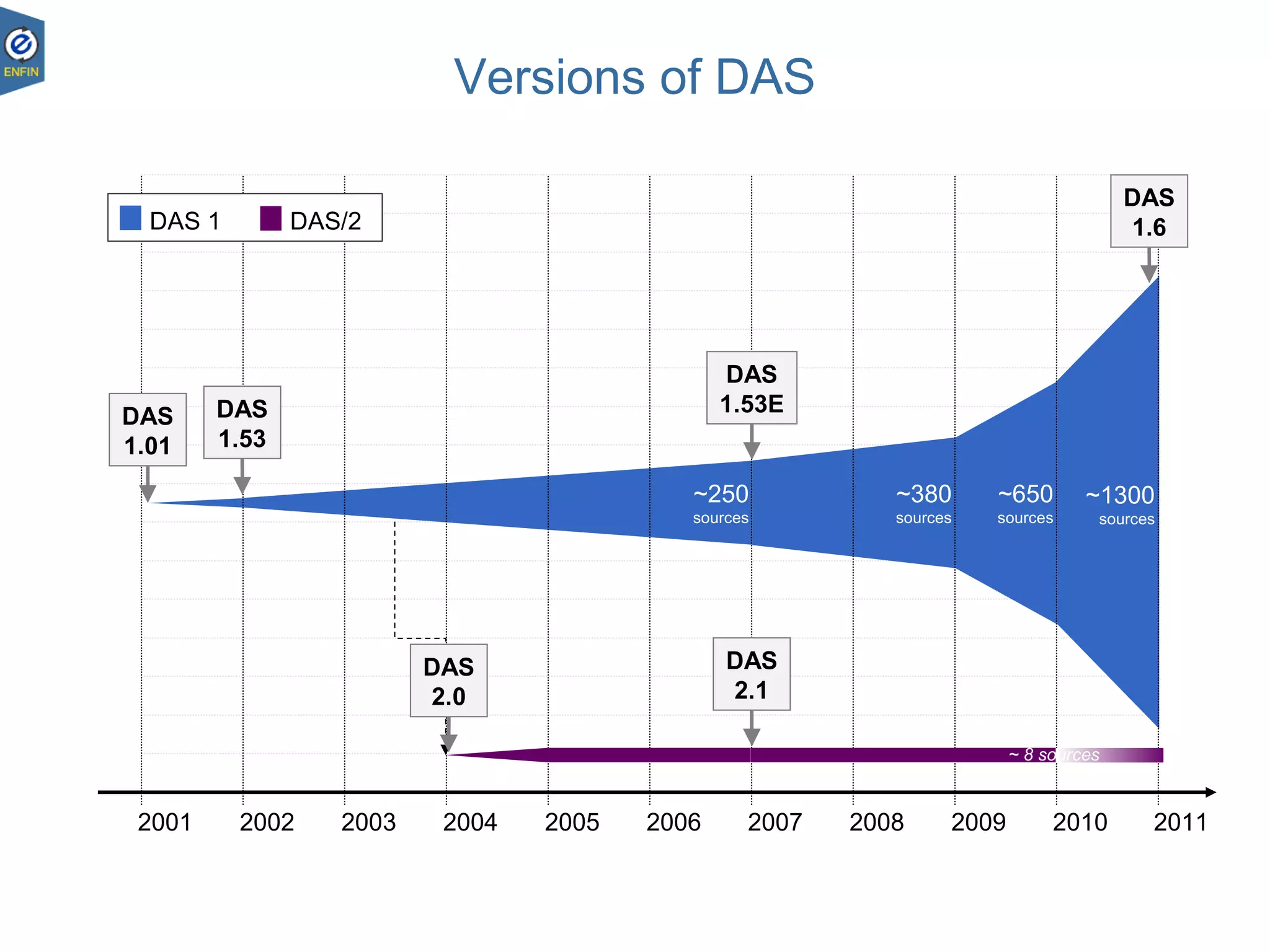
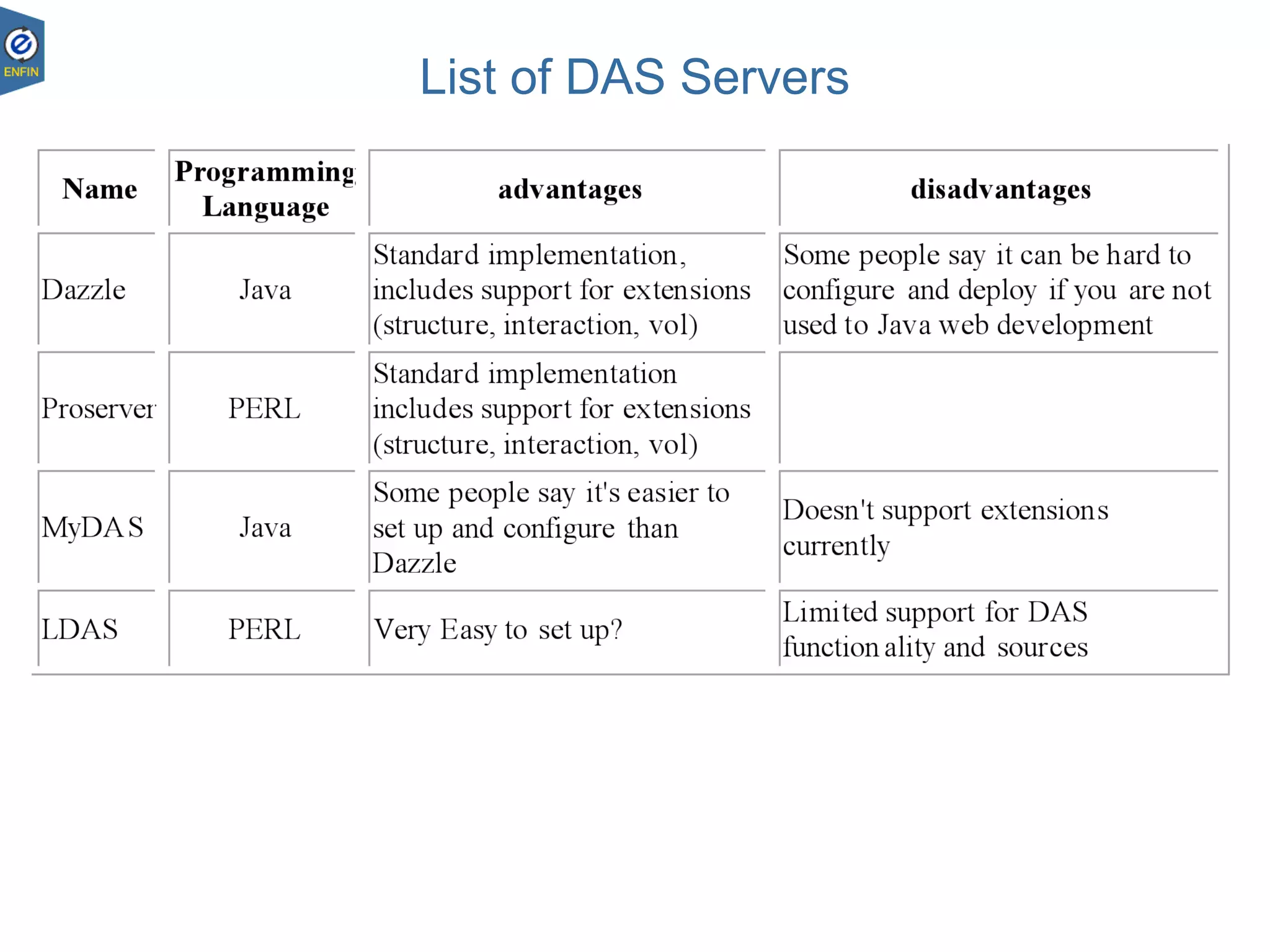
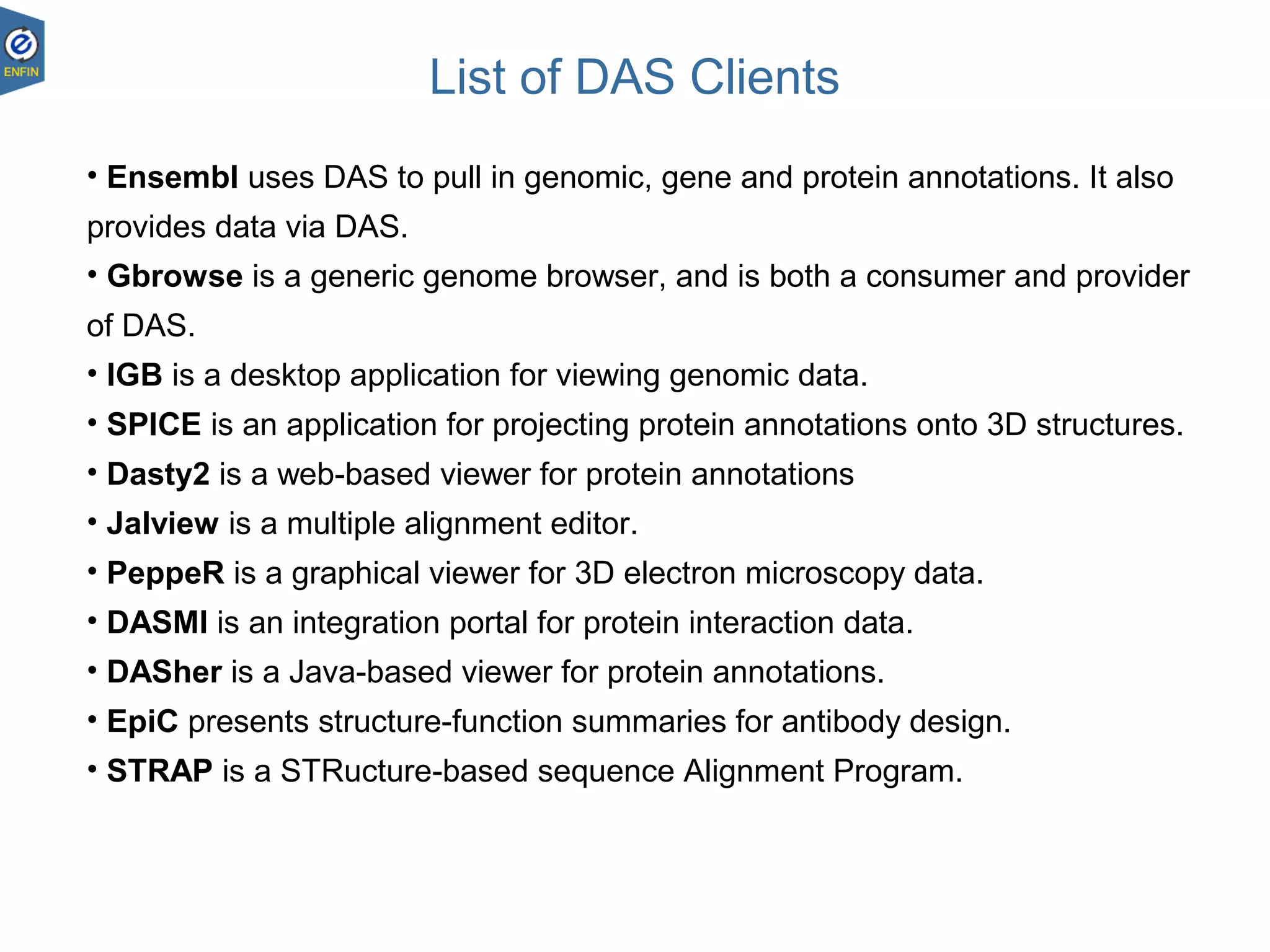
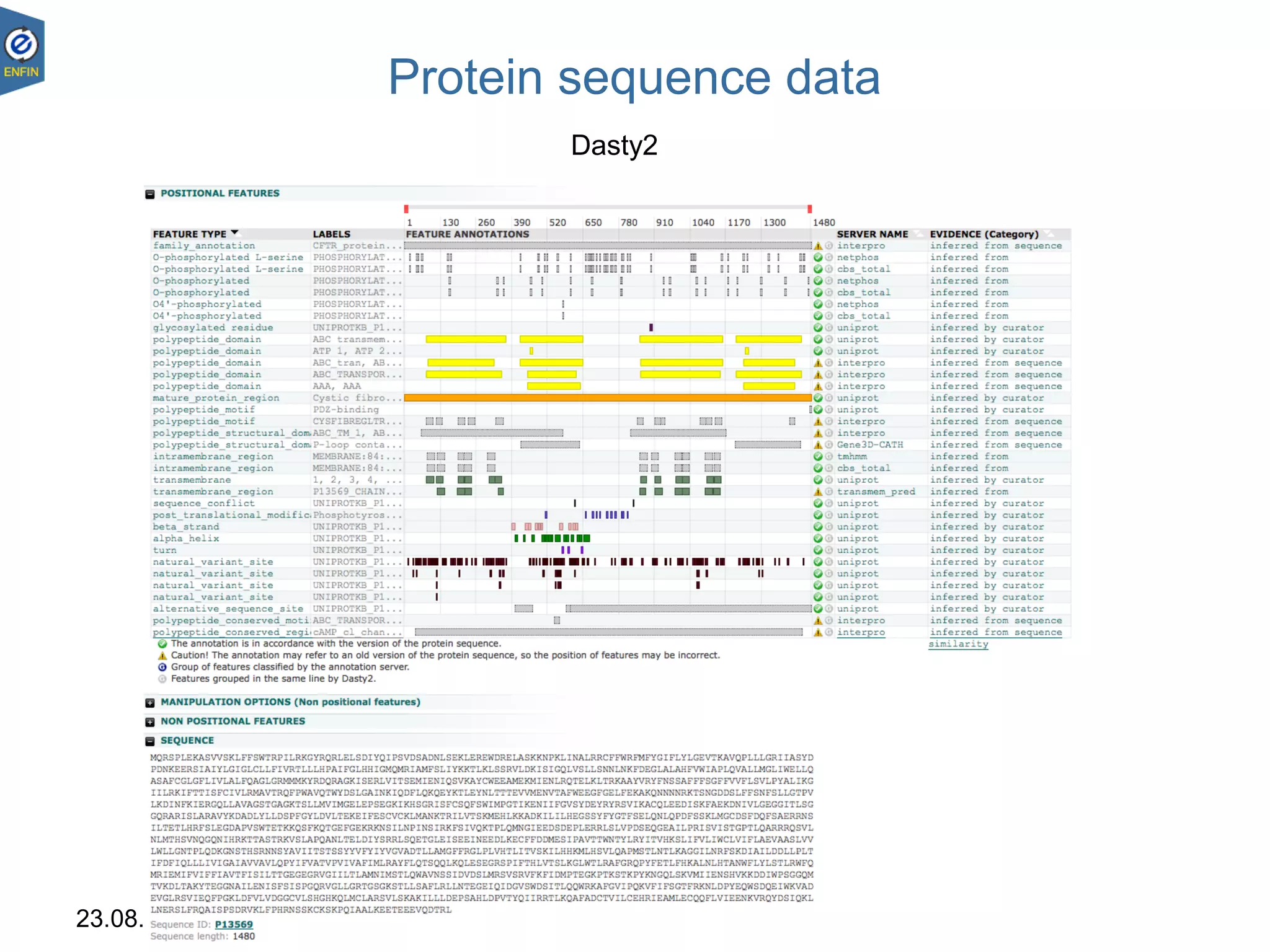
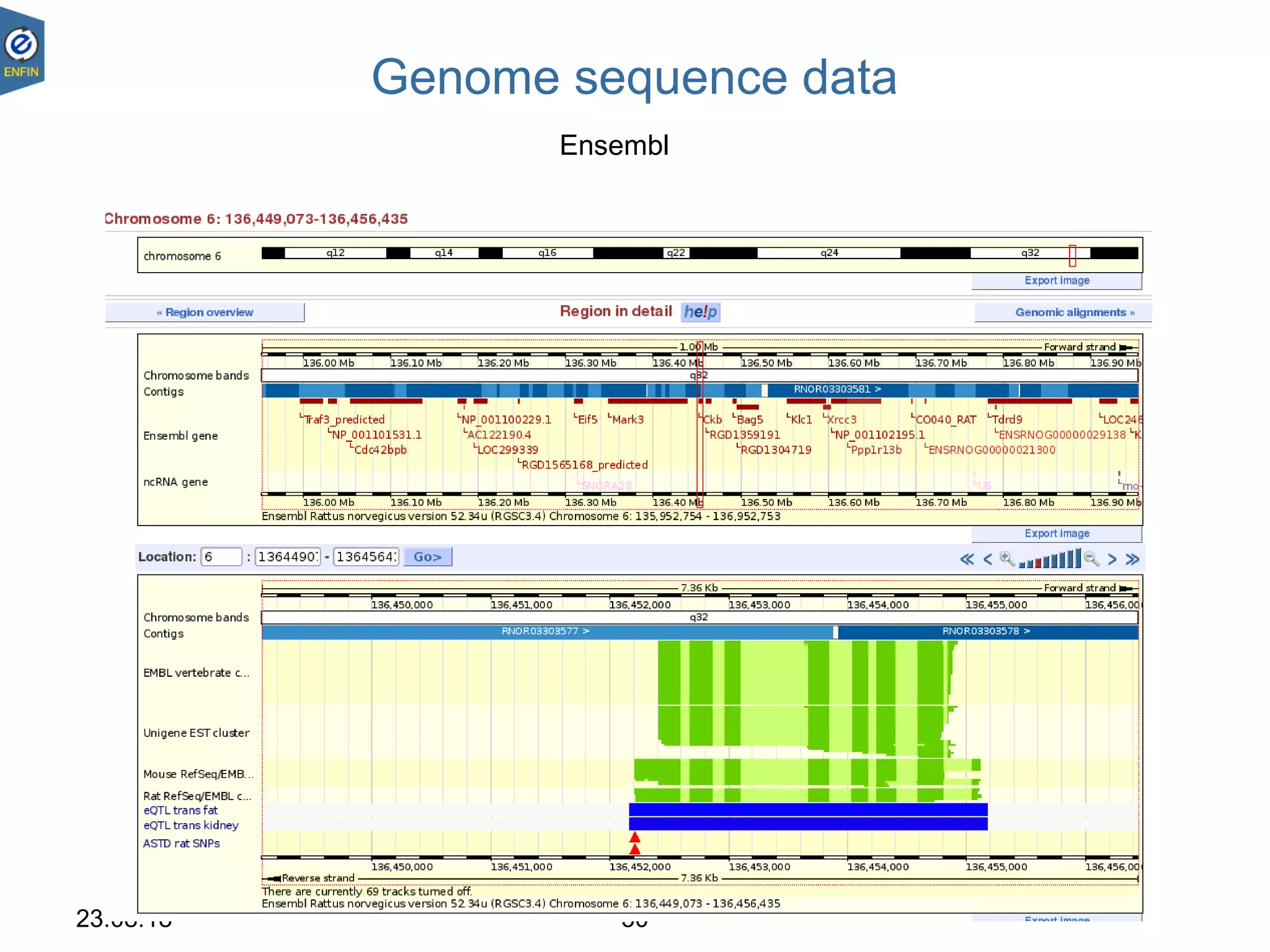
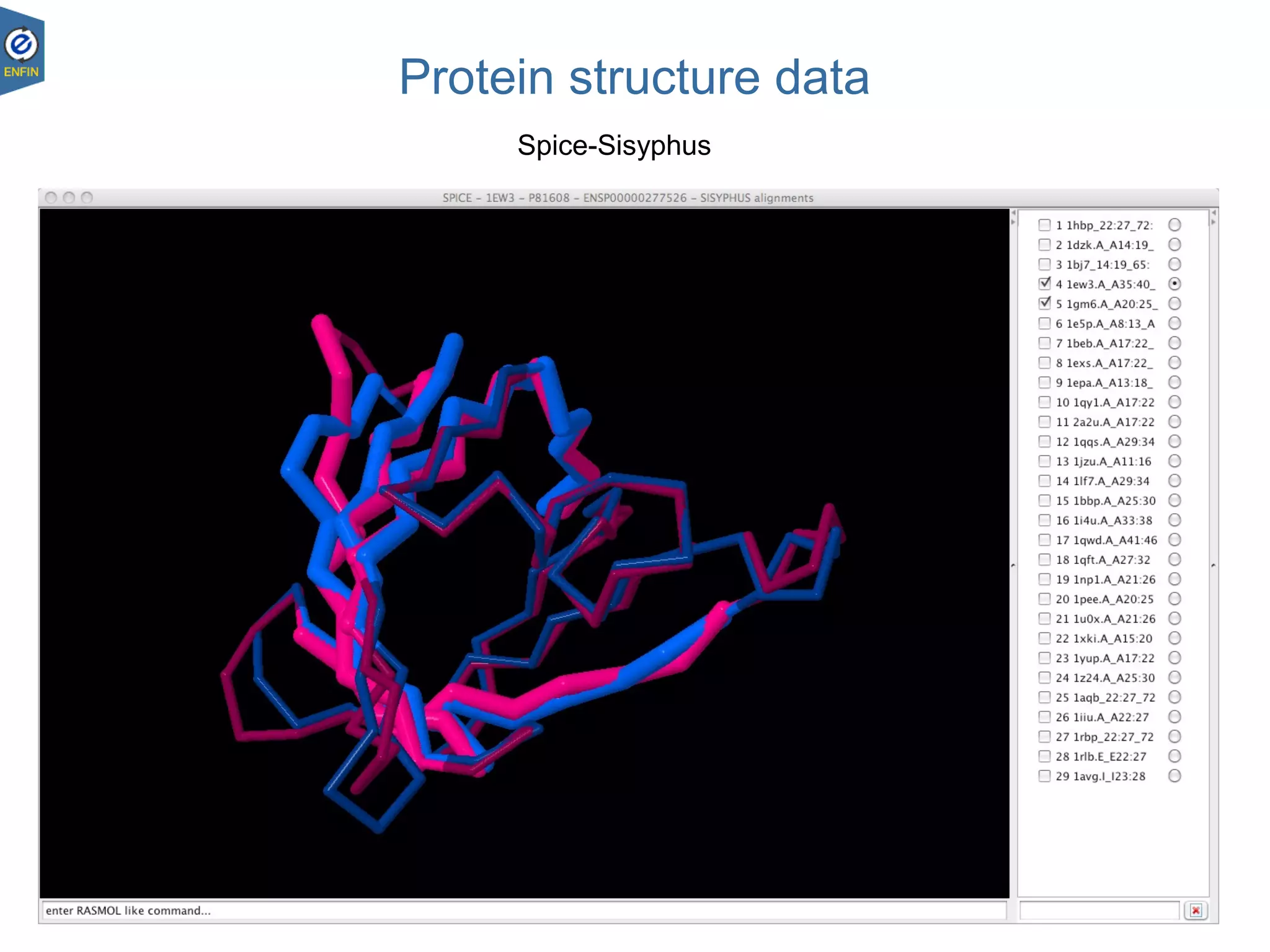
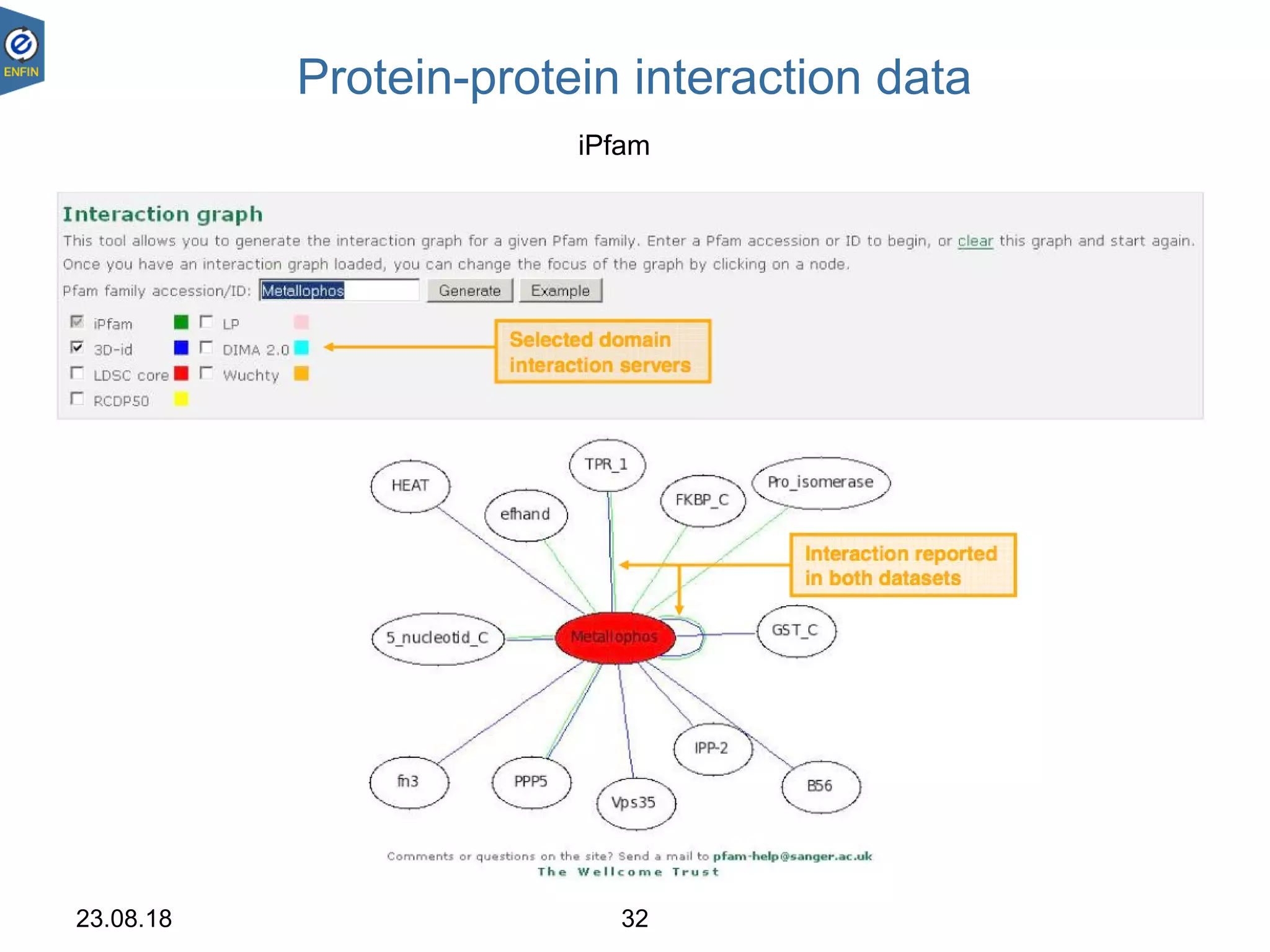
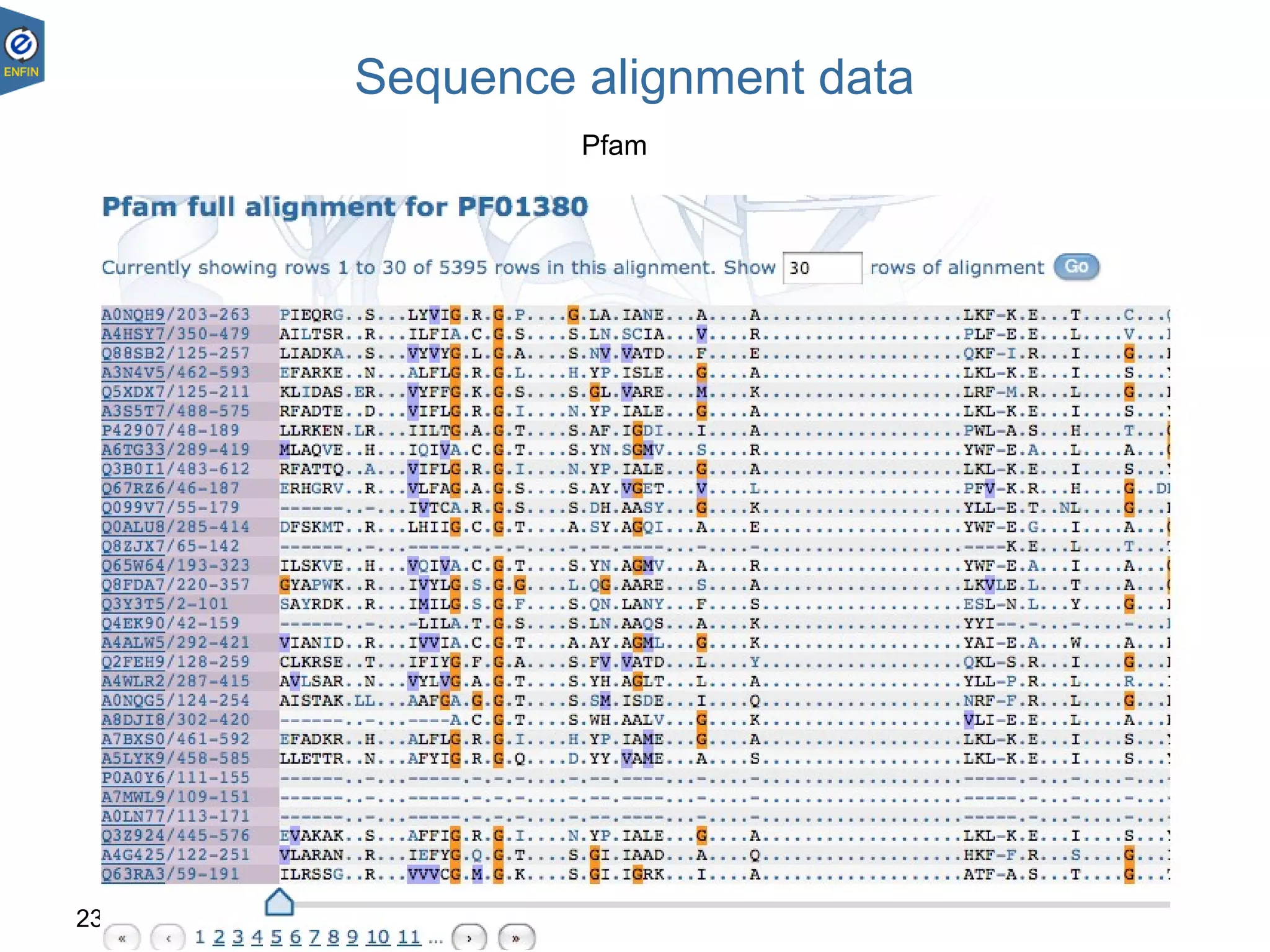
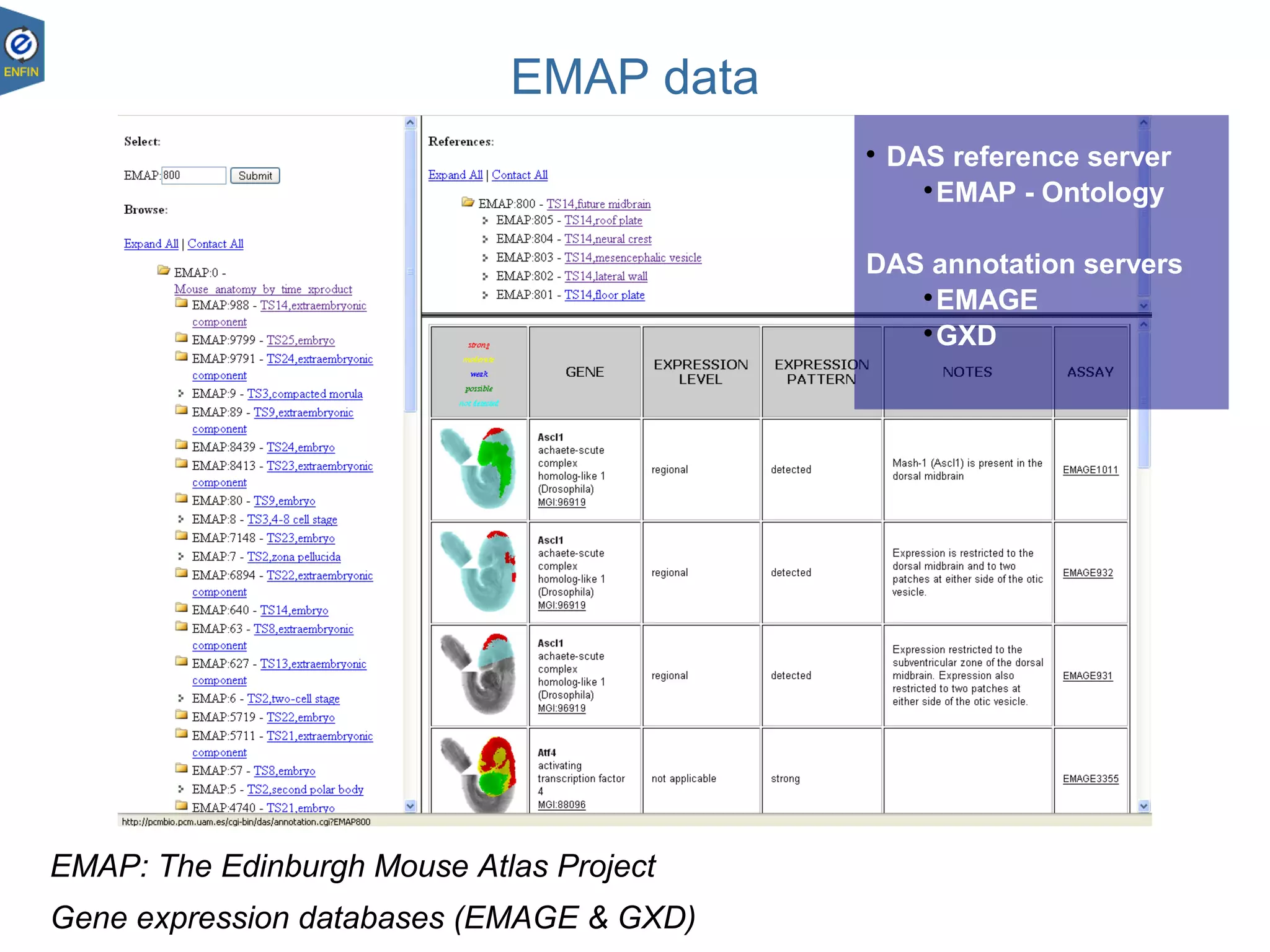
This document provides an overview of the Distributed Annotation System (DAS), which is a network of biological data sources that allows for uniform access to distributed repositories of biological data through a standardized client-server protocol. It describes what DAS is, how clients can query servers using commands and a lightweight XML data format, examples of DAS clients and servers, and documentation resources for the DAS specification and protocol versions. The goal of DAS is to provide an integration platform and service-oriented architecture for federated access to annotation data across different sources.