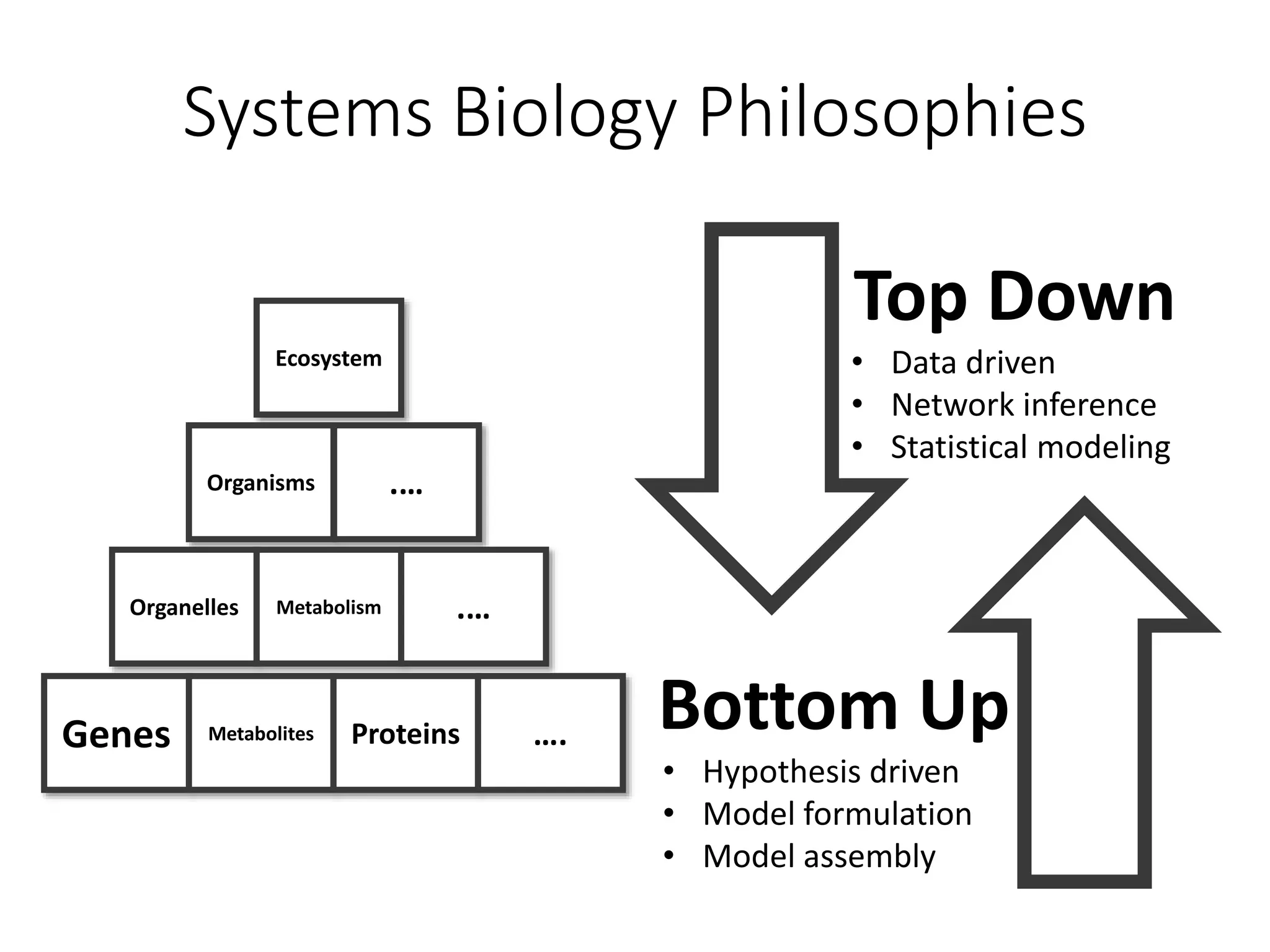
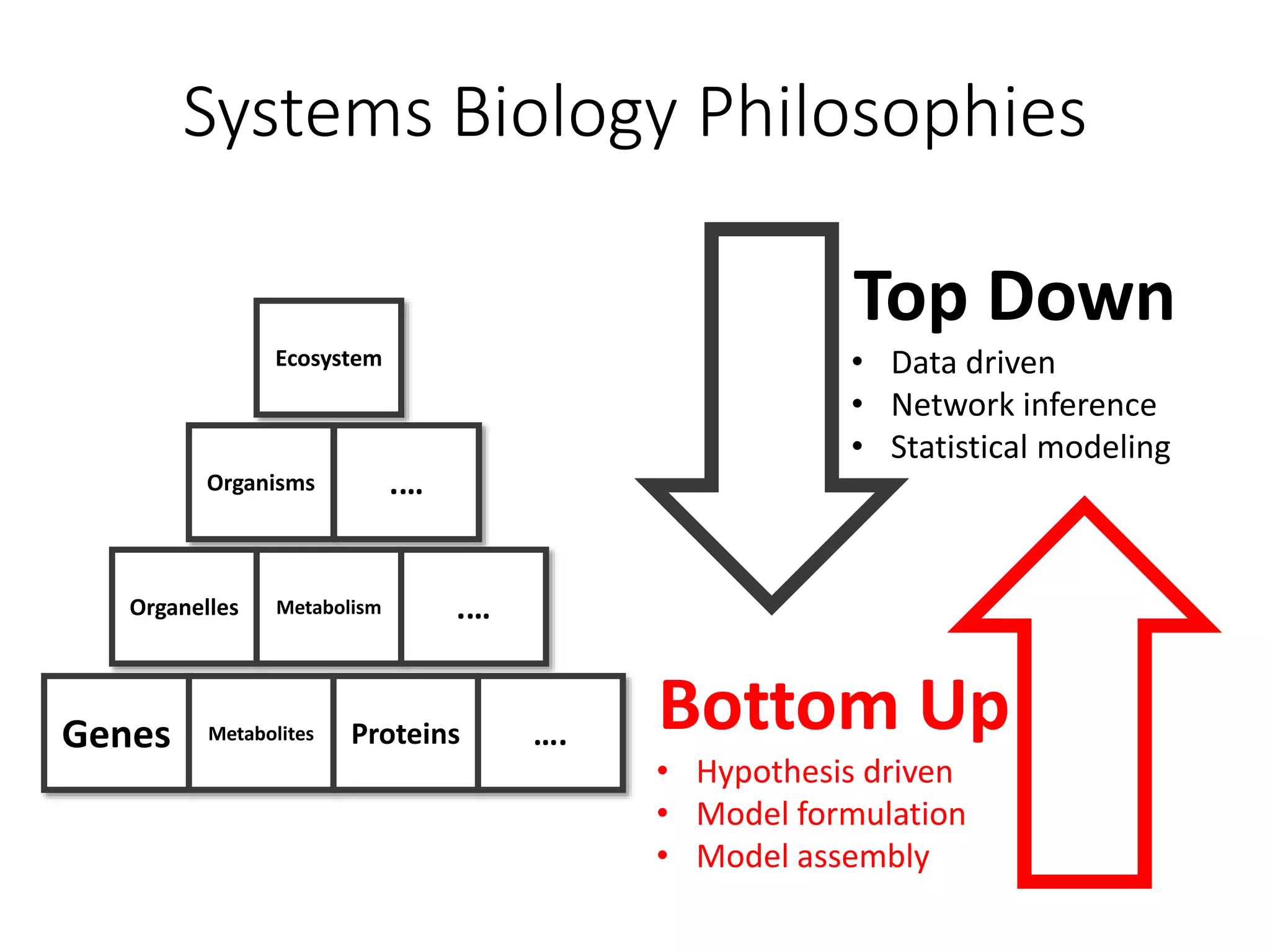
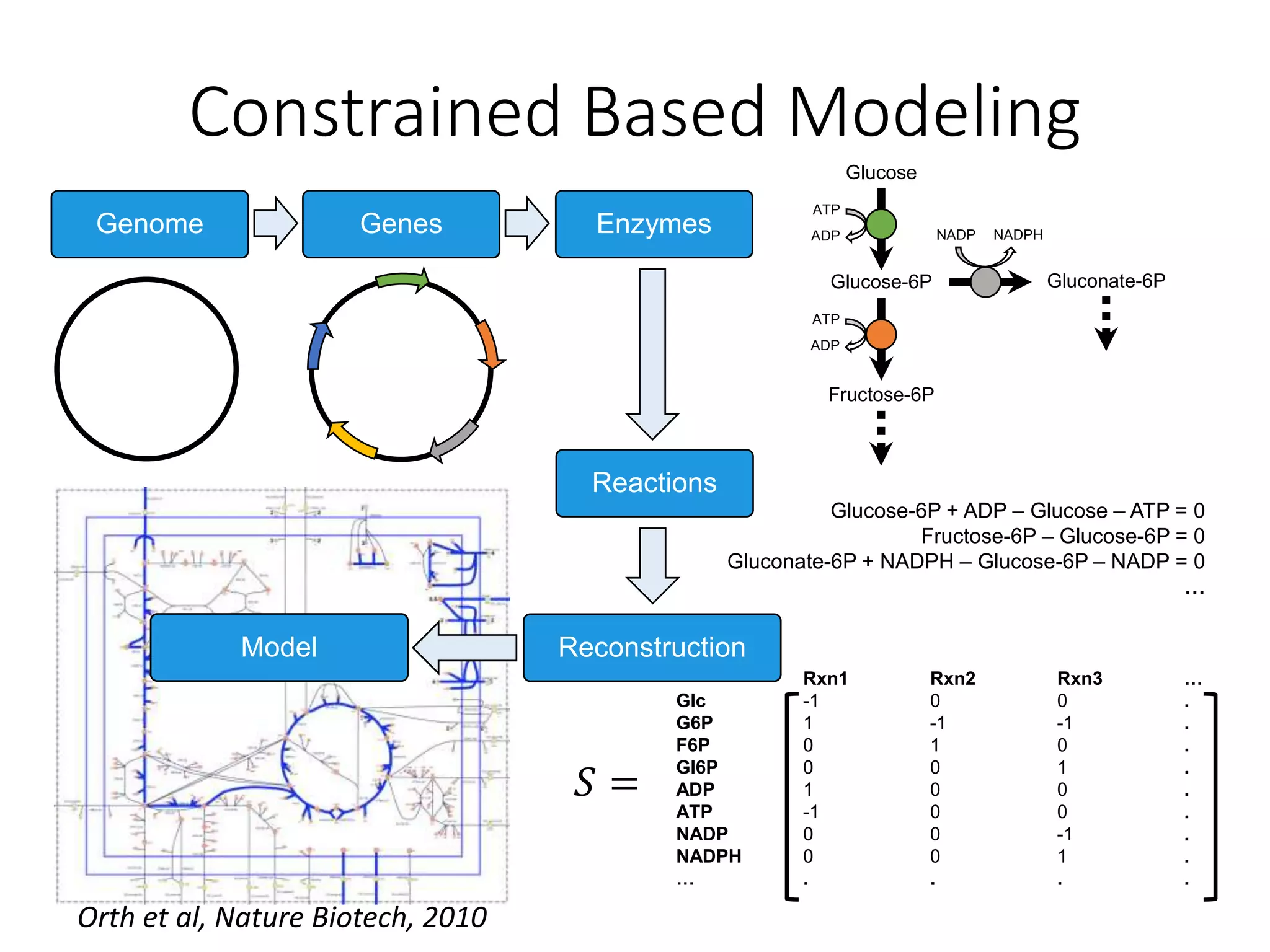
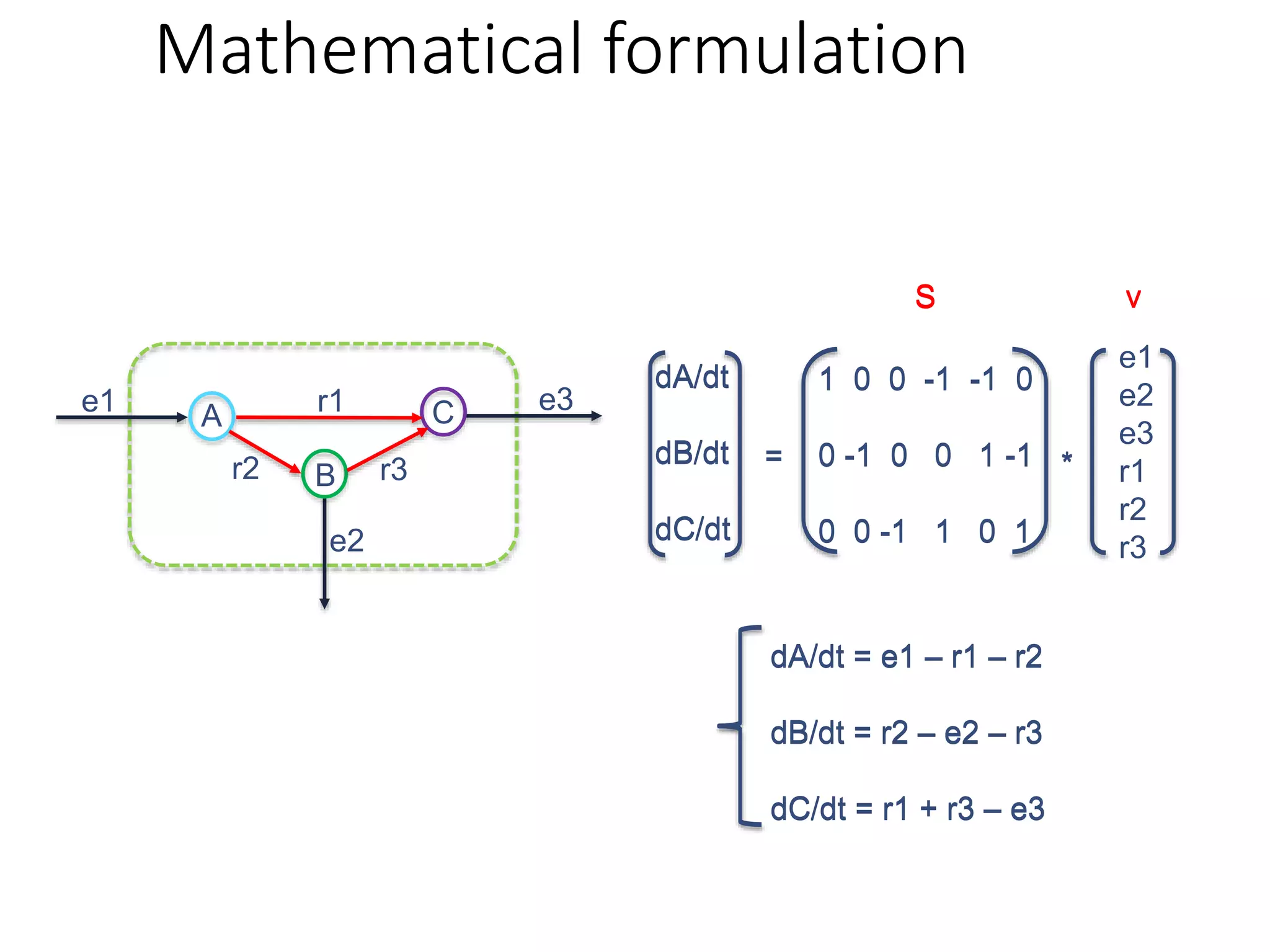
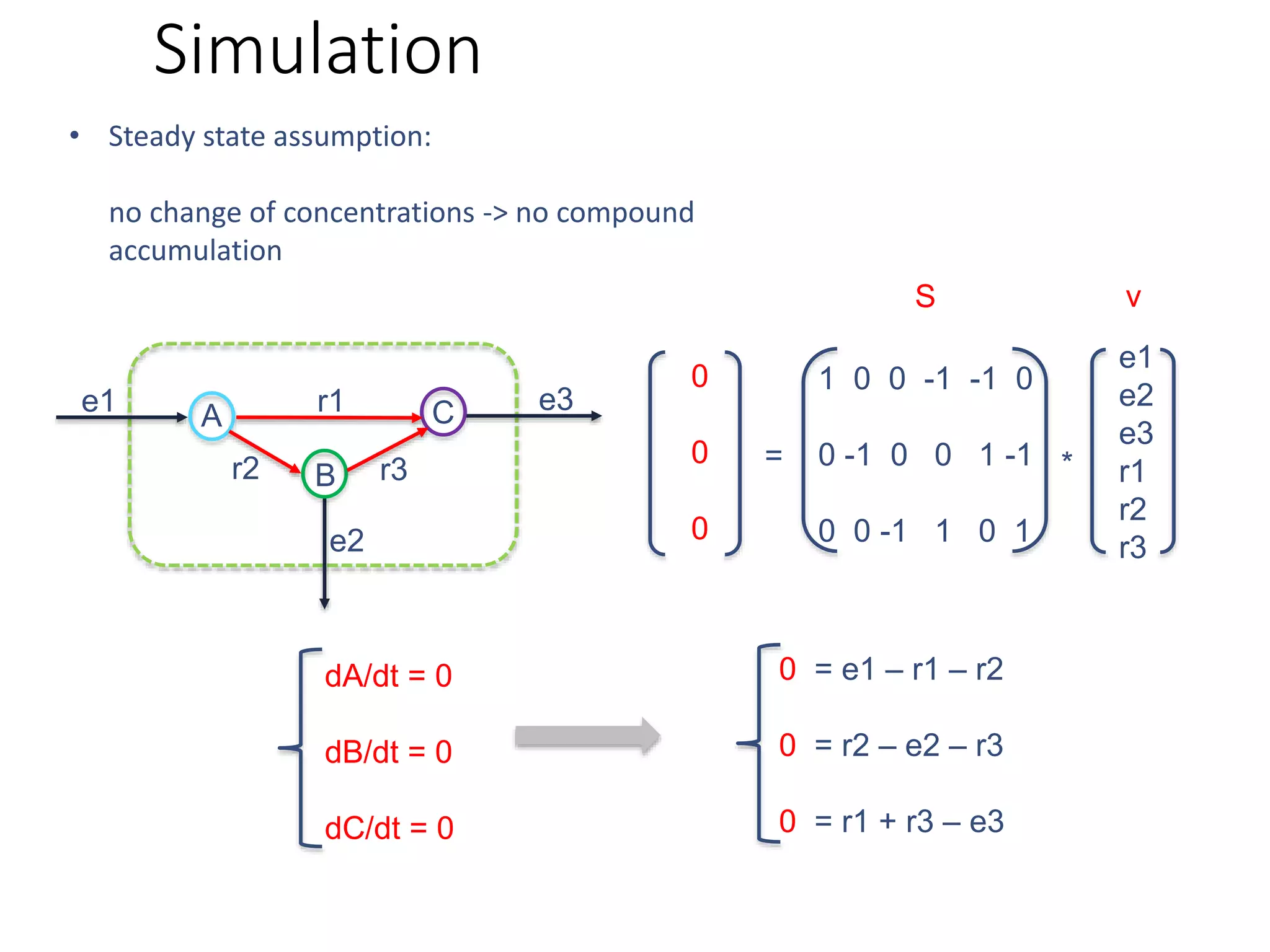
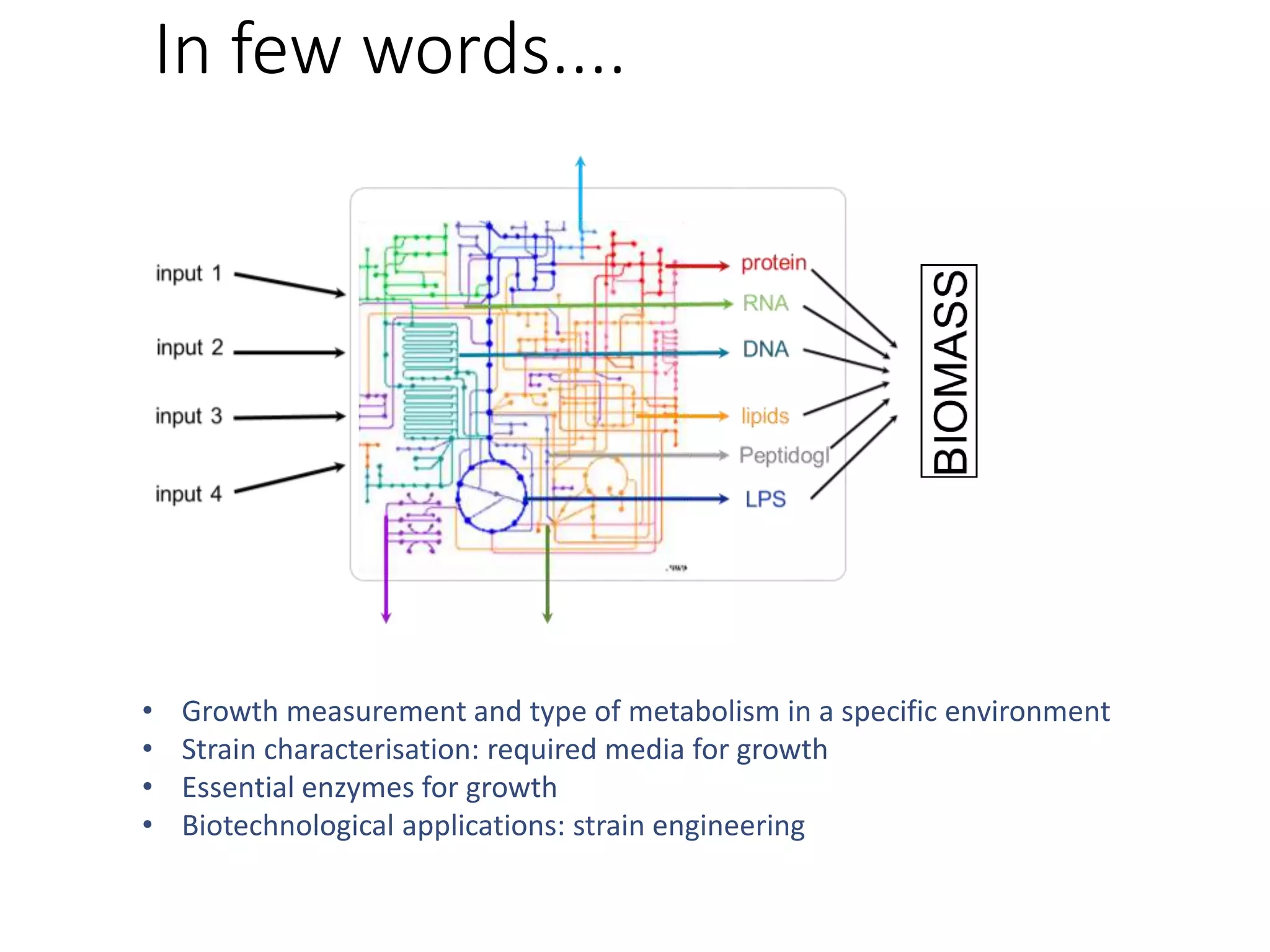
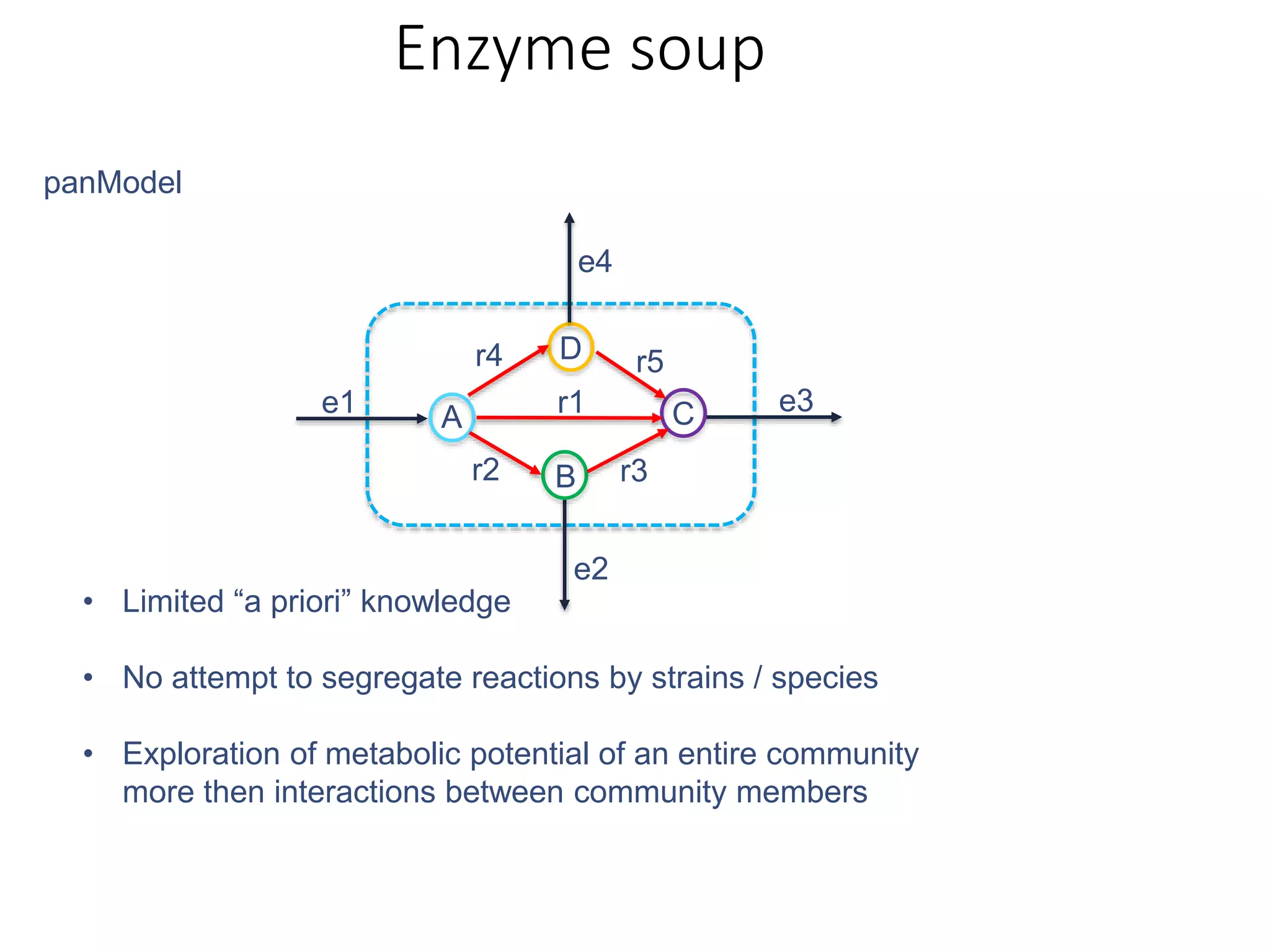
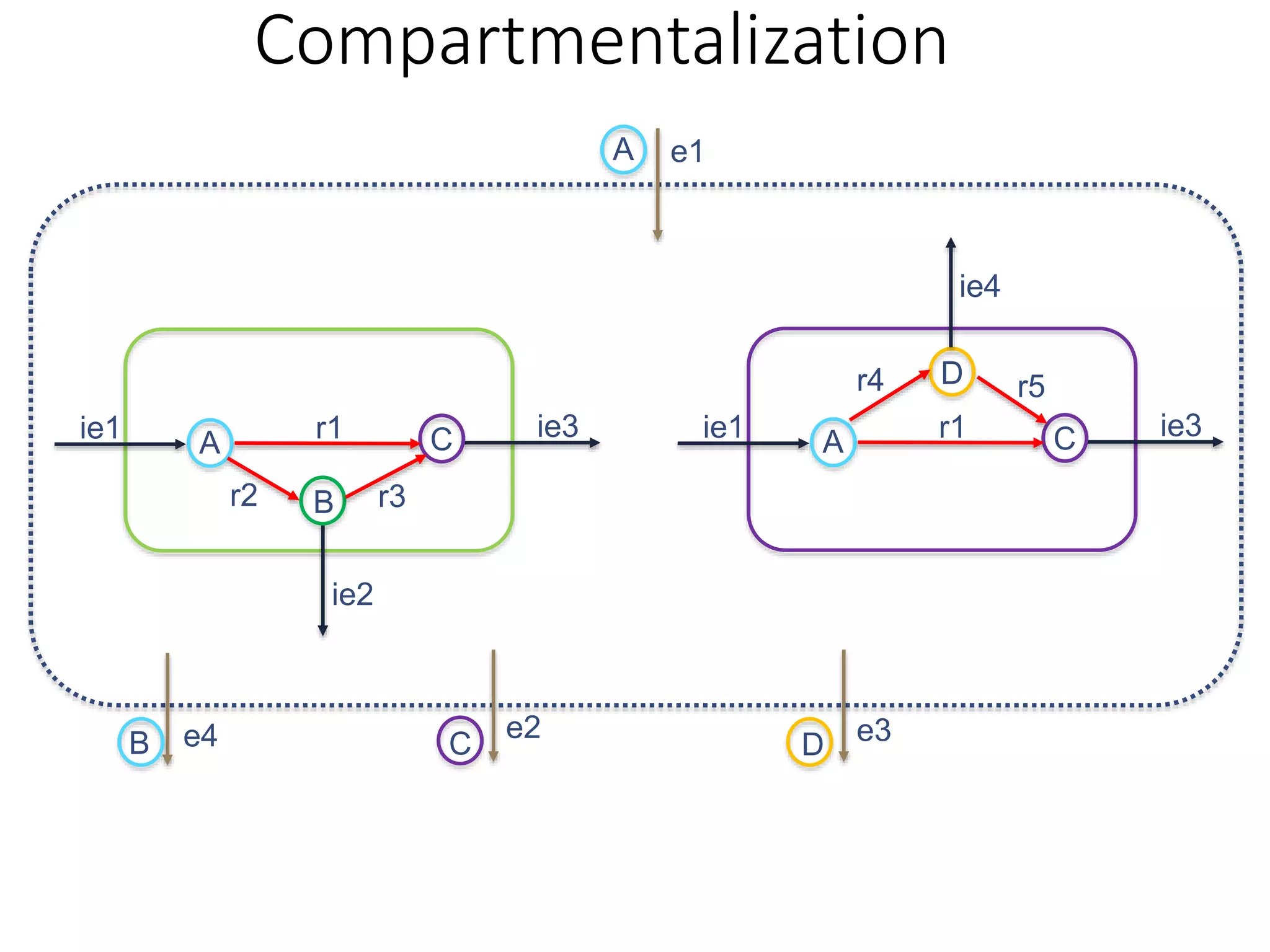
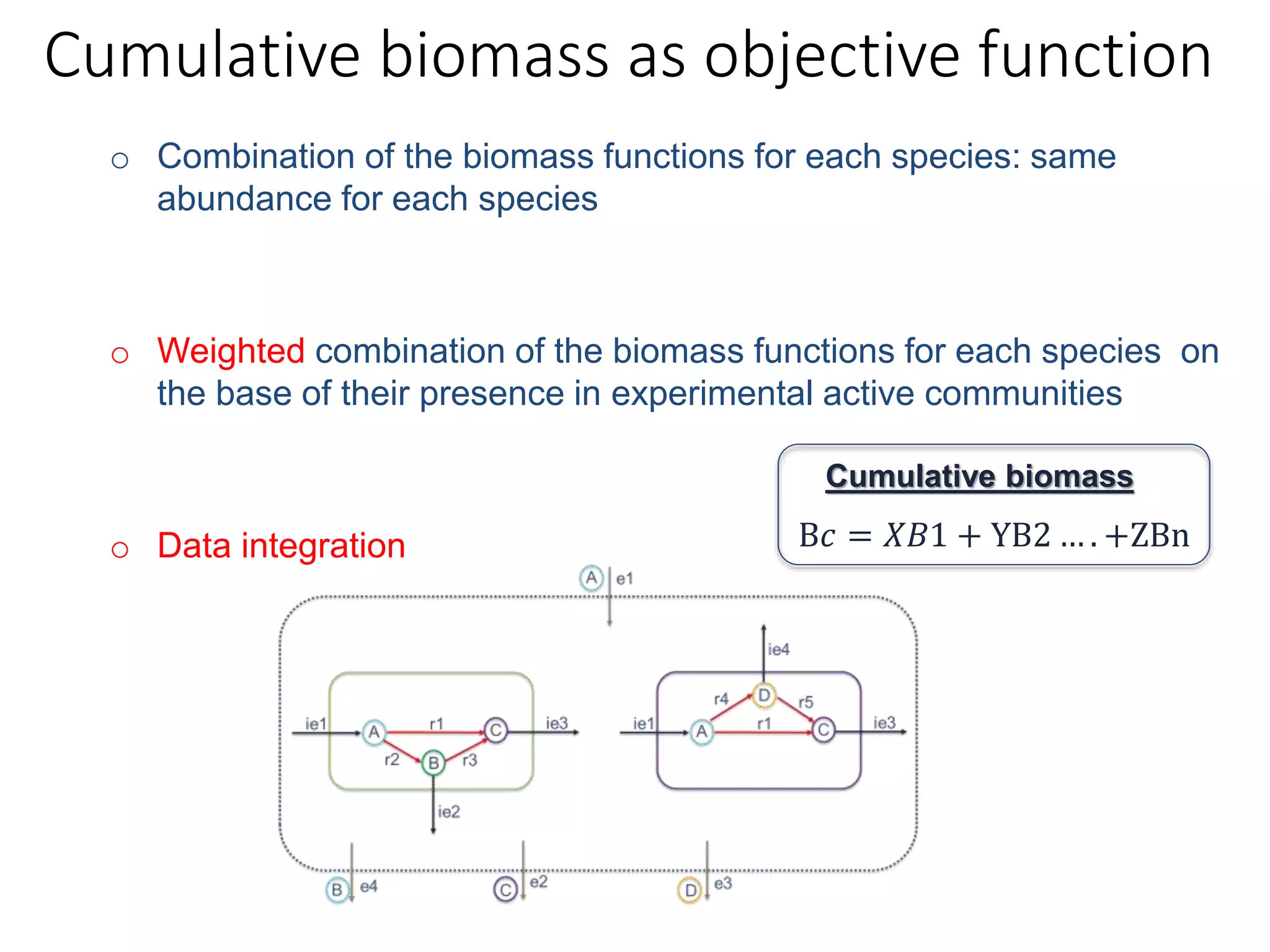
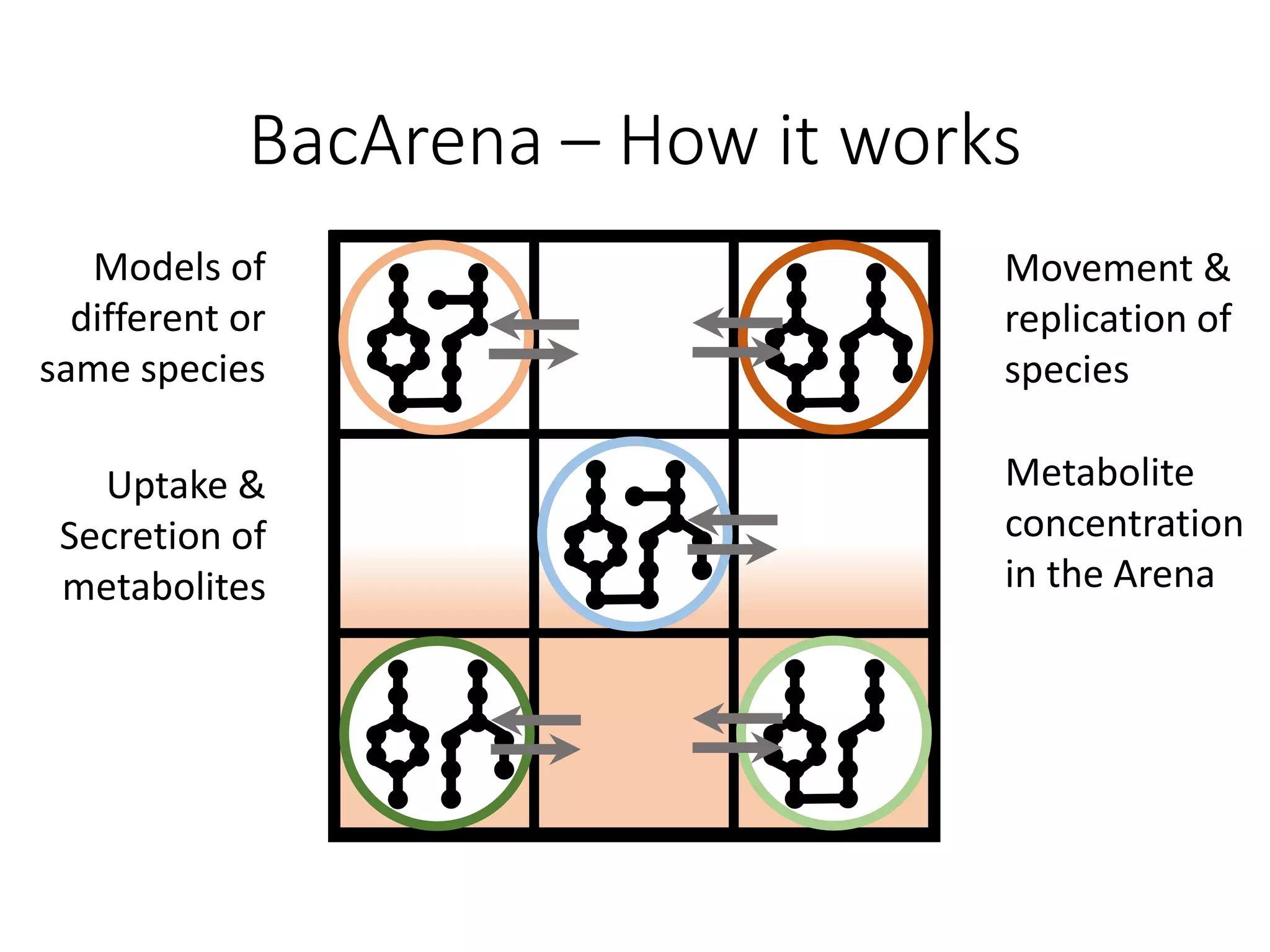
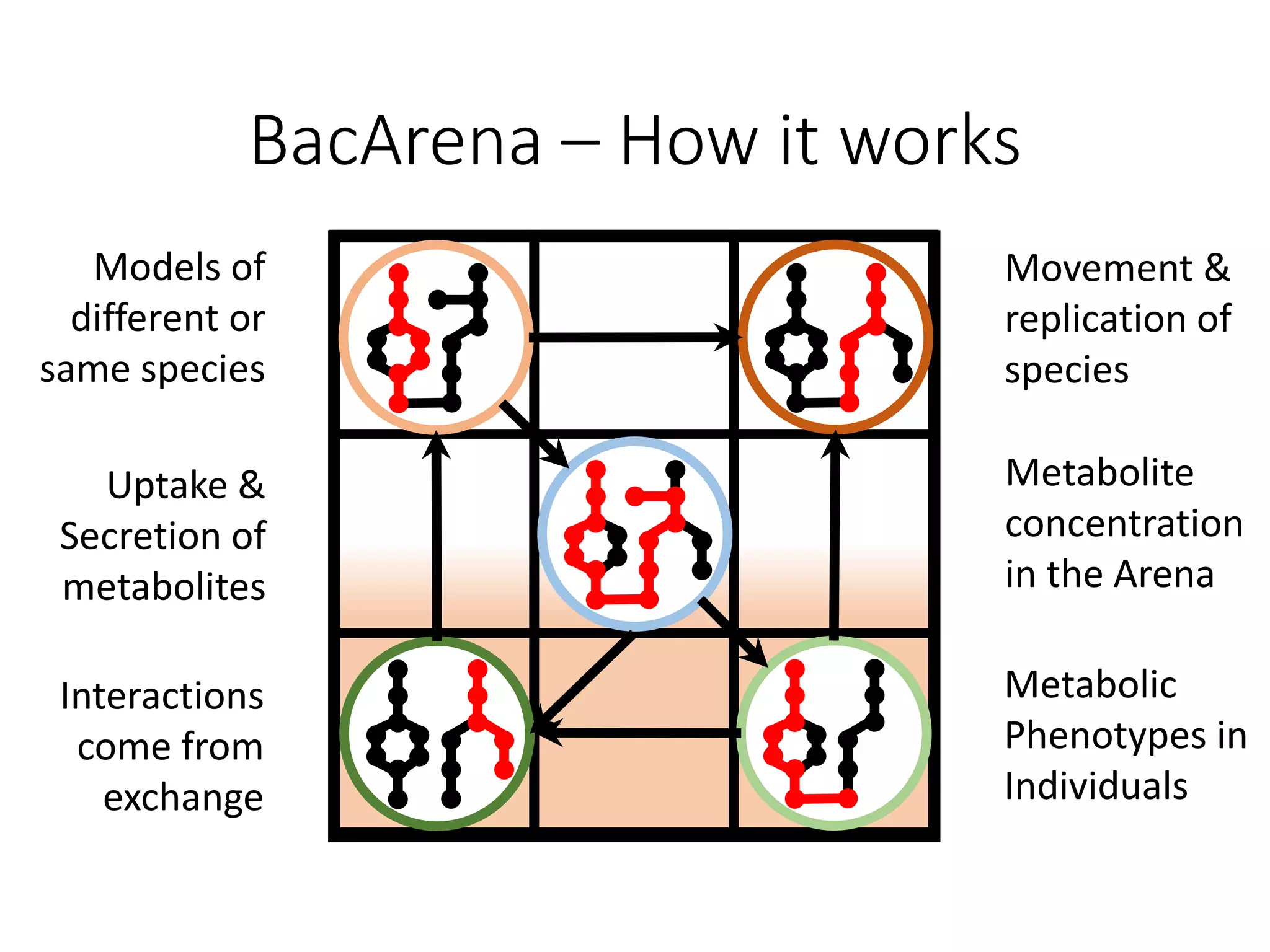
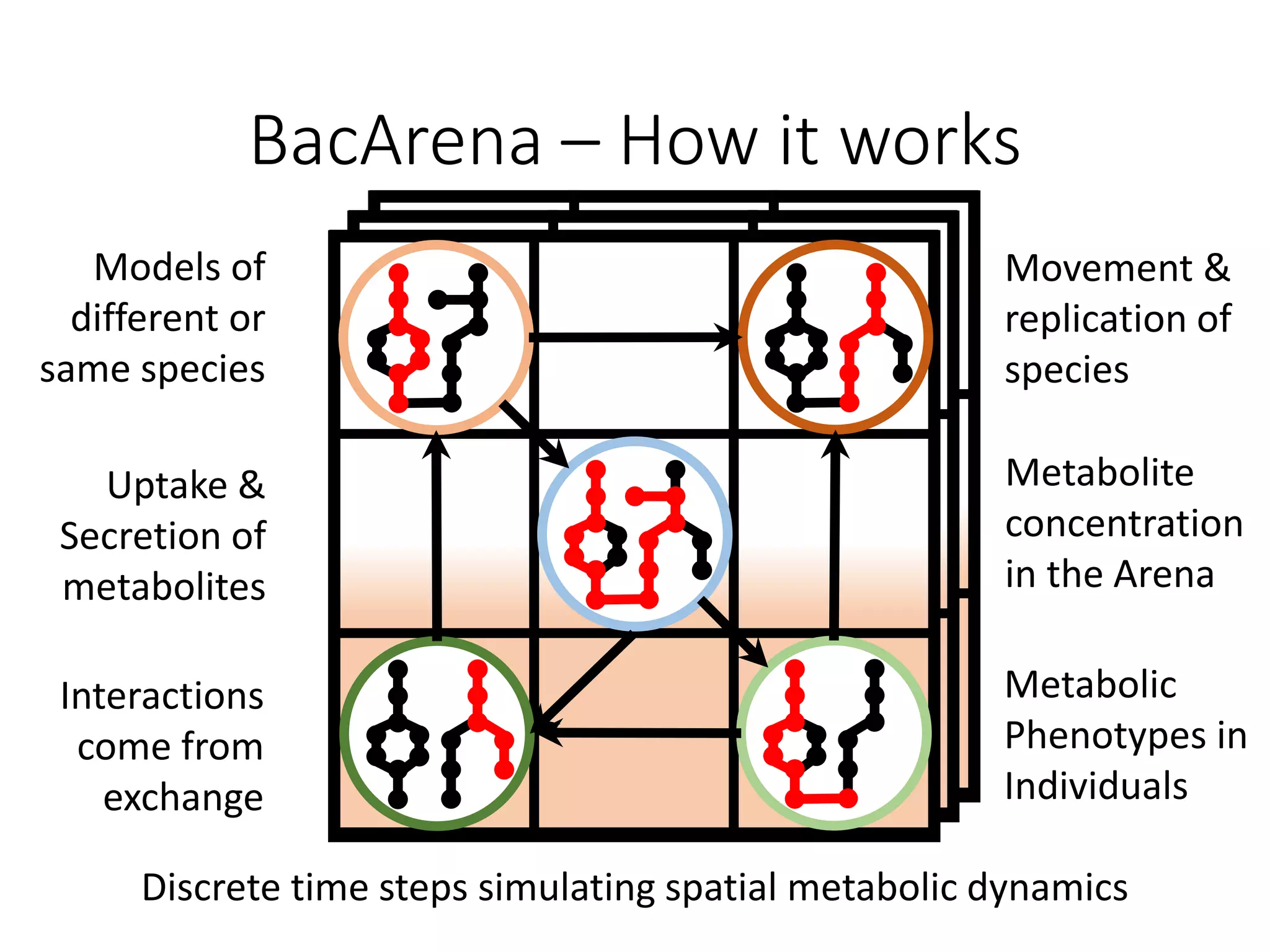
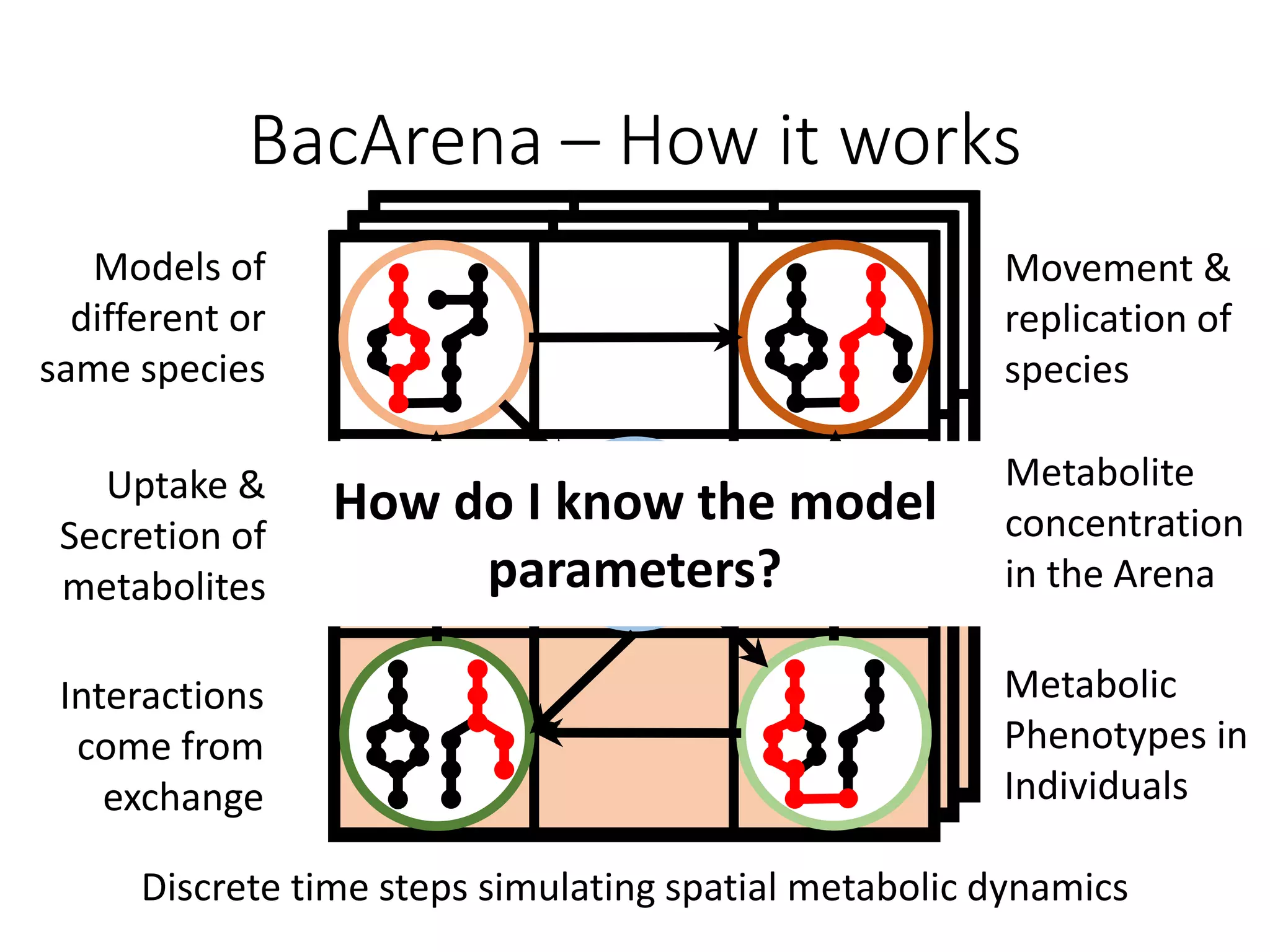
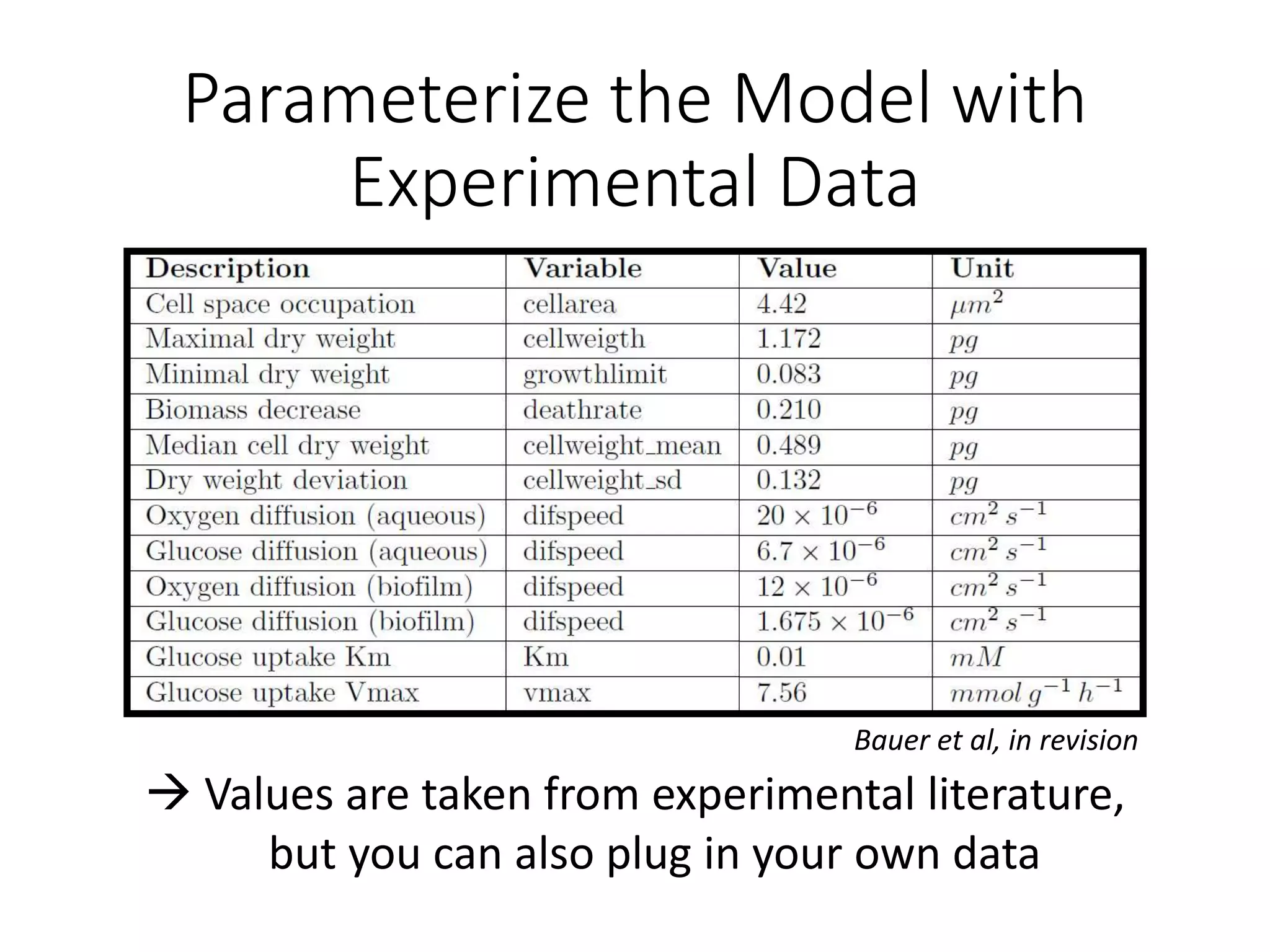
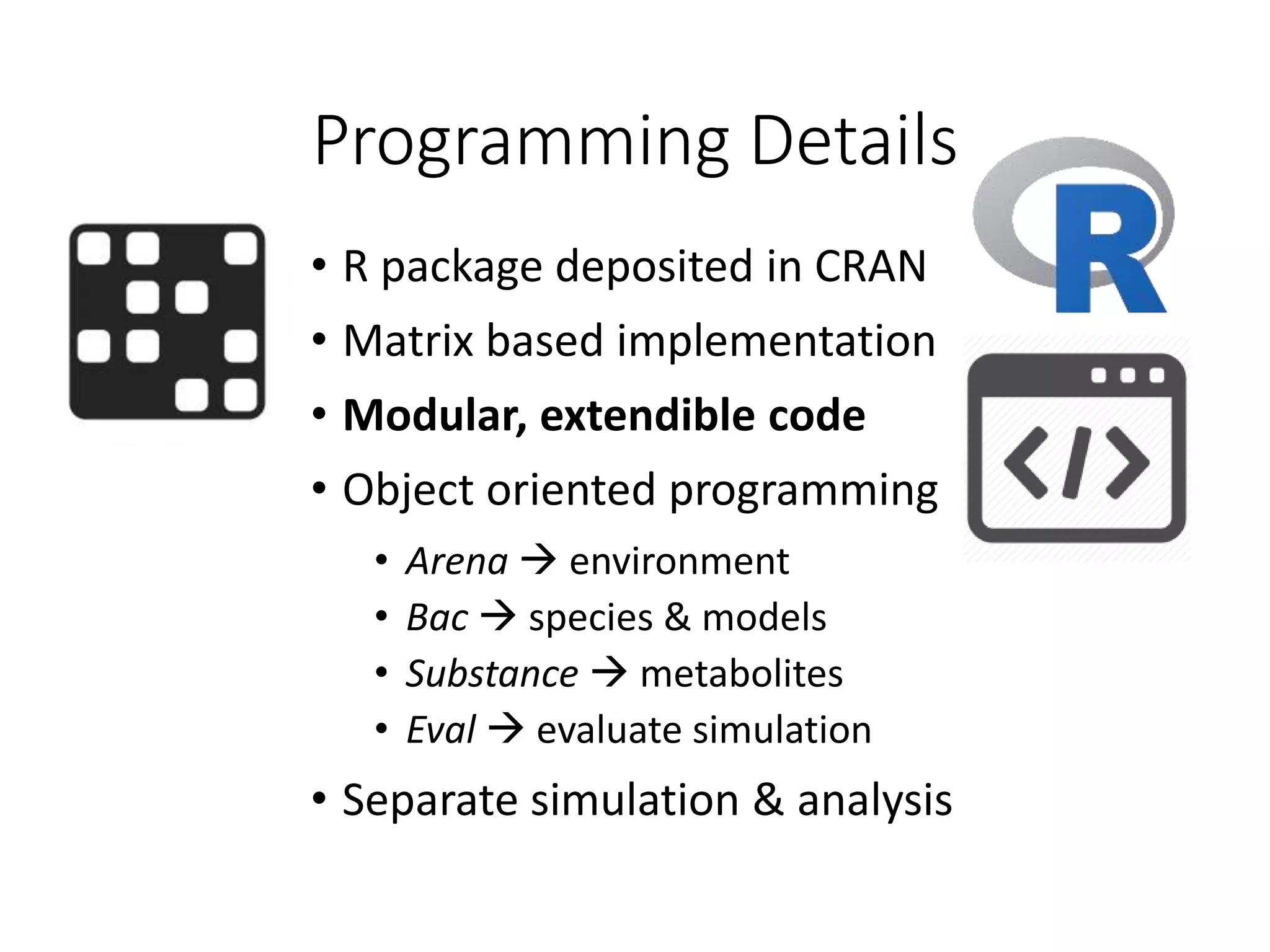
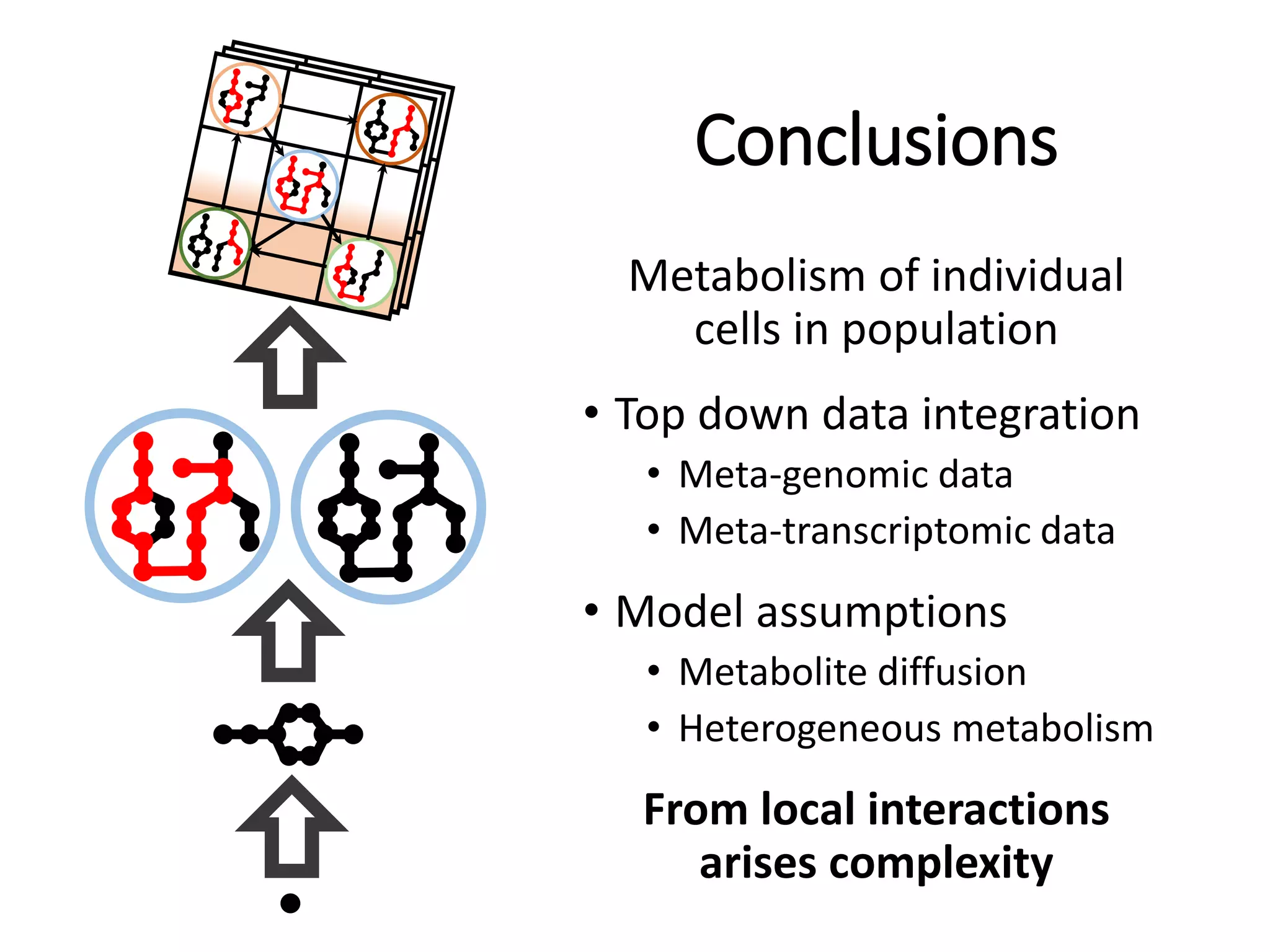
The document outlines a workshop on community modeling led by Federico Baldini and Eugen Bauer, focusing on systems biology and the significance of interactions among biological components. It introduces a tool named Bacarena for simulating metabolic dynamics within microbial communities, employing both constrained and agent-based models. The workshop includes discussions on theoretical frameworks, practical applications, and programming details for model implementation.