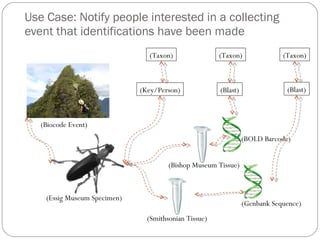
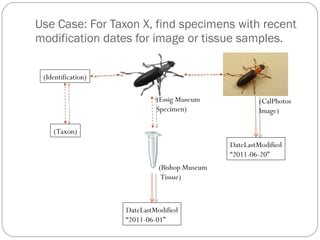
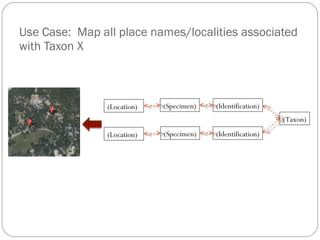
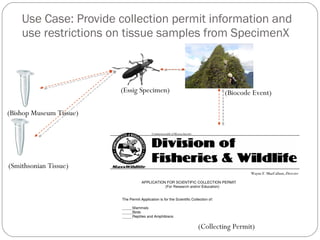
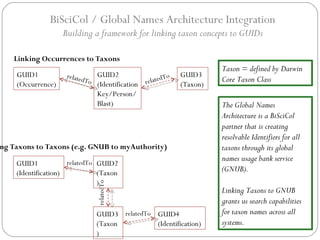
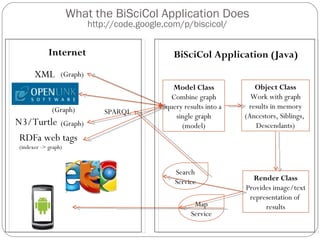
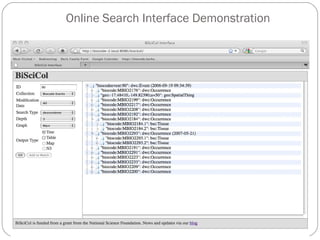
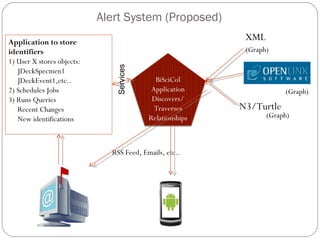
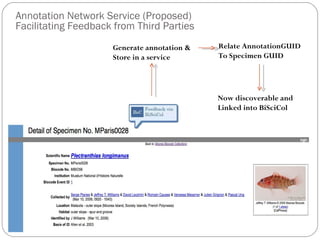
The document discusses the Biscicol project, an initiative aimed at creating a tagging and tracking infrastructure for biological science collections, funded by the National Science Foundation between 2010 and 2014. It highlights the use of globally unique identifiers (GUIDs) to track specimens and proposes various use cases that showcase the significance and functionality of the Biscicol application. The project involves collaboration among multiple institutions including UC Berkeley, Colorado University, and the Smithsonian Institution, and emphasizes the integration of linked data and the Global Names Architecture.