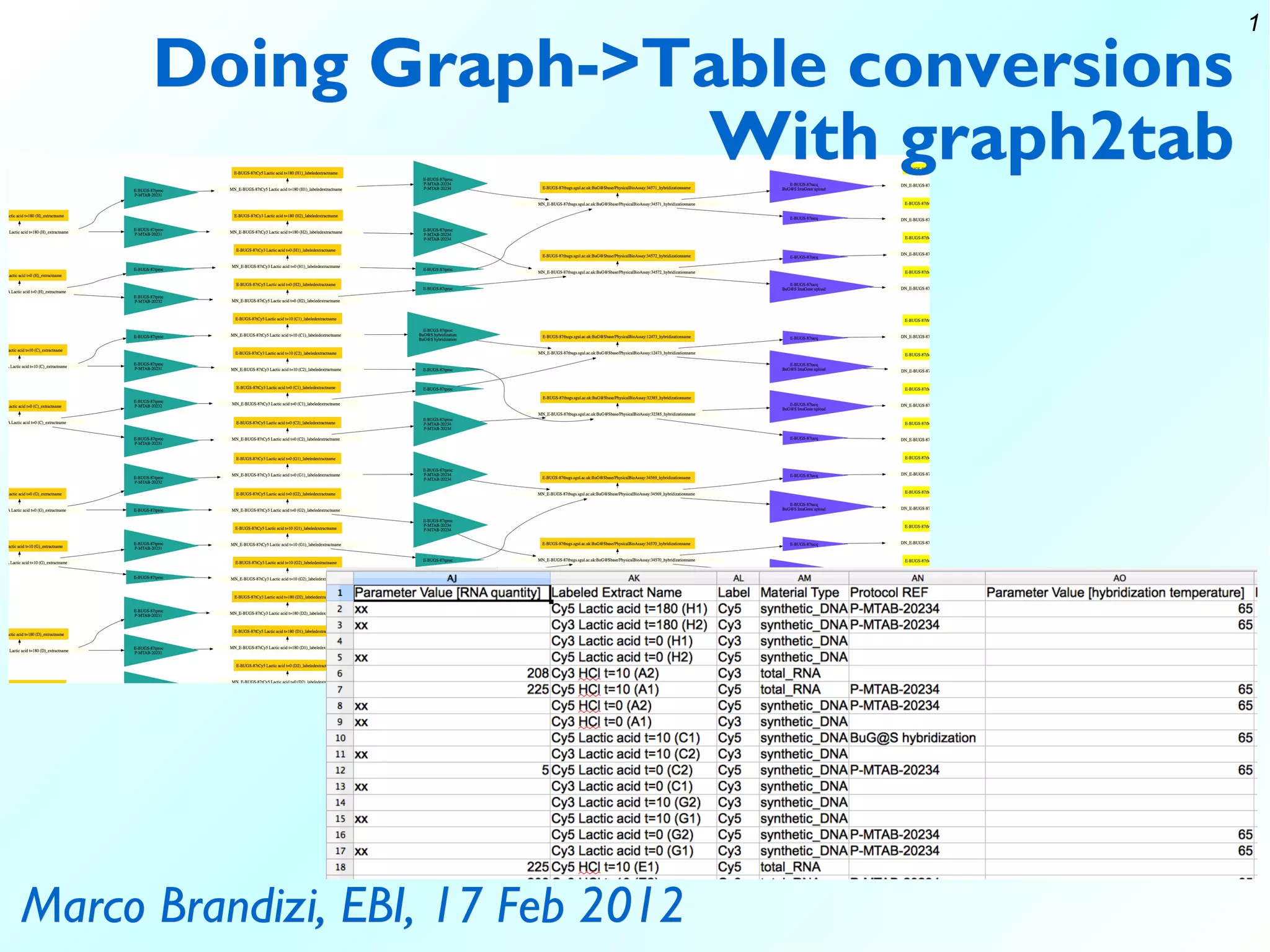
The document discusses the functionalities of graph2tab, a tool designed for converting graph data into tabular formats such as ISA-tab and Magetab, with a focus on math and technical principles including graph theory and optimization processes. It outlines the complexities involved in finding optimal path sets to convert graphs effectively while addressing computational limitations in biology-related data interpretation. Furthermore, the document highlights various use cases and acknowledges contributions from team members and collaborators involved in the development of this tool.


















![20
Conceptually simple, but...
Map<String, Integer> TYPE_ORDER =
new HashMap<String, Integer> ()
{{
put ( "Source Name", 0 );
put ( "Sample Name", 1 );
put ( "Extract Name", 2 );
put ( "Labeled Extract Name", 3 );
put ( "Assay Name", 4 );
put ( "Data File Name", 5 );
}};
Ah-Ah! Now I know what toAh-Ah! Now I know what to
choose!choose!
Ah-Ah! Now I know what toAh-Ah! Now I know what to
choose!choose!
+interface Node
extends Comparable<Node>
{
getInputs (): SortedSet<Node>
getOutputs (): SortedSet<Node>
getTabValues (): TabValueGroup[]
getType (): String
getOrder (): int
}](https://image.slidesharecdn.com/graph2tabebi20120217-171003215930/75/graph2tab-a-library-to-convert-experimental-workflow-graphs-into-tabular-formats-20-2048.jpg)



![24
Representing Node Attributes
TabValueGroup (
header = "Organism"
value = "Mus-mus"
tail = ( TabValueGroup (
header = "Term Source REF"
value = "NCBI-Tax"
tail = ( TabValueGroup (
header = "Term Accession"
value = "123"
))
))
)
StructuredTable (
header = "Organism"
rows = "R. Norvegicus"
tail = []
)
StructuredTable (
header = "Organism"
rows = "R. Norvegicus", "Mus-mus"
tail = ( StructuredTable (
header = "Term Source REF"
rows = "", "", "NCBI-Tax"
tail = ( StructuredTable (
header = "Term Accession"
rows = "", "", "123"
))
))
)
+ =](https://image.slidesharecdn.com/graph2tabebi20120217-171003215930/75/graph2tab-a-library-to-convert-experimental-workflow-graphs-into-tabular-formats-24-2048.jpg)







