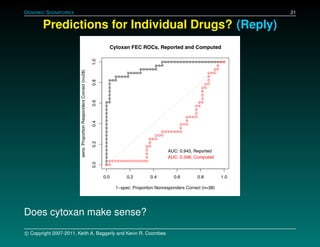
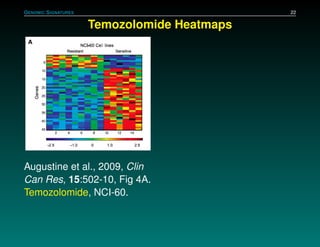
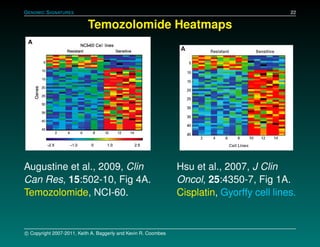
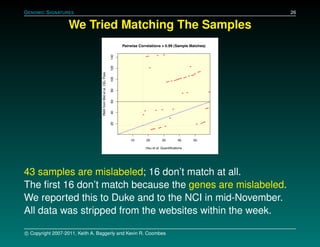
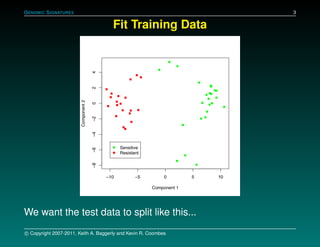
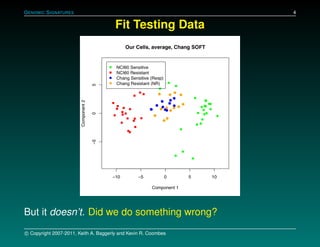
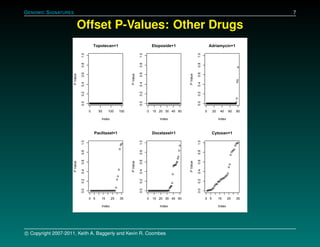
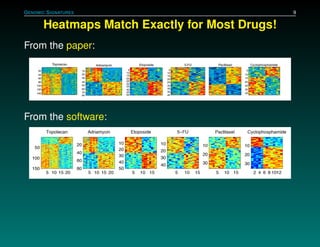
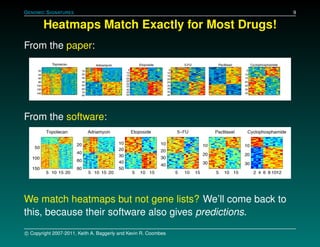
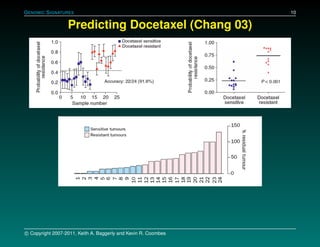
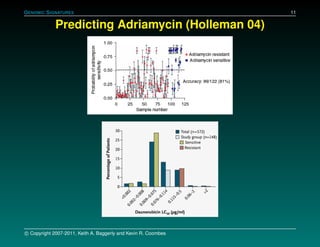
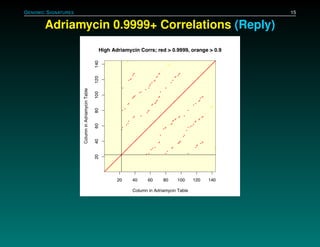
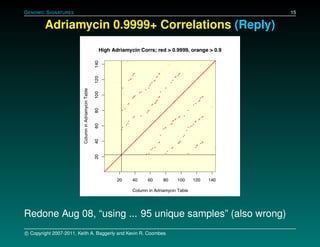
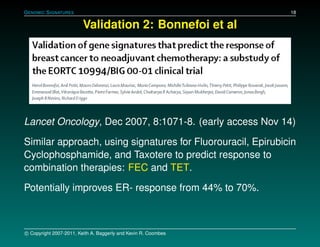
The document discusses several case studies involving predicting cancer patient response to chemotherapy using gene expression signatures. It summarizes that researchers claimed they could predict drug response using microarray data from cell lines, but upon external analysis several issues were found, such as gene lists not matching and poor performance on test data. The document examines multiple studies that claimed to validate the approach but found inconsistencies in results and undocumented changes to data and methods. It emphasizes the importance of reproducibility in high-throughput studies and encourages readers to examine the raw data themselves.






![G ENOMIC S IGNATURES 6
Their List and Ours
> temp <- cbind(
sort(rownames(pottiUpdated)[fuRows]),
sort(rownames(pottiUpdated)[
fuTQNorm@p.values <= fuCut]);
> colnames(temp) <- c("Theirs", "Ours");
> temp
Theirs Ours
...
[3,] "1881_at" "1882_g_at"
[4,] "31321_at" "31322_at"
[5,] "31725_s_at" "31726_at"
[6,] "32307_r_at" "32308_r_at"
...
c Copyright 2007-2011, Keith A. Baggerly and Kevin R. Coombes](https://image.slidesharecdn.com/baggerlycse11-1-110503202731-phpapp02/85/Baggerly-presentation-from-CSE-9-320.jpg)








![G ENOMIC S IGNATURES 20
How Are Results Combined?
Potti et al predict response to TFAC, Bonnefoi et al to TET
and FEC. Let P() indicate prob sensitive. The rules used are
as follows.
P (T F AC) = P (T )+P (F )+P (A)+P (C)−P (T )P (F )P (A)P (C).
P (ET ) = max[P (E), P (T )].](https://image.slidesharecdn.com/baggerlycse11-1-110503202731-phpapp02/85/Baggerly-presentation-from-CSE-28-320.jpg)
![G ENOMIC S IGNATURES 20
How Are Results Combined?
Potti et al predict response to TFAC, Bonnefoi et al to TET
and FEC. Let P() indicate prob sensitive. The rules used are
as follows.
P (T F AC) = P (T )+P (F )+P (A)+P (C)−P (T )P (F )P (A)P (C).
P (ET ) = max[P (E), P (T )].
5 1
P (F EC) = [P (F ) + P (E) + P (C)] − .
8 4](https://image.slidesharecdn.com/baggerlycse11-1-110503202731-phpapp02/85/Baggerly-presentation-from-CSE-29-320.jpg)
![G ENOMIC S IGNATURES 20
How Are Results Combined?
Potti et al predict response to TFAC, Bonnefoi et al to TET
and FEC. Let P() indicate prob sensitive. The rules used are
as follows.
P (T F AC) = P (T )+P (F )+P (A)+P (C)−P (T )P (F )P (A)P (C).
P (ET ) = max[P (E), P (T )].
5 1
P (F EC) = [P (F ) + P (E) + P (C)] − .
8 4
Each rule is different.
c Copyright 2007-2011, Keith A. Baggerly and Kevin R. Coombes](https://image.slidesharecdn.com/baggerlycse11-1-110503202731-phpapp02/85/Baggerly-presentation-from-CSE-30-320.jpg)