More Related Content
Similar to Senior Thesis Poster
Similar to Senior Thesis Poster (20)
Senior Thesis Poster
- 1. © File copyright Colin Purrington. You
may use for making your poster, of
course, but please do not plagiarize,
adapt, or put on your own site. Also,
do not upload this file, even if
modified, to third-party file-sharing
sites such as doctoc.com. If you have
insatiable need to post a template onto
your own site, search the internet for a
different template to steal. File
downloaded from http://
colinpurrington.com/tips/academic/
posterdesign.
Background:
Astroviruses
are
a
major
cause
of
diarrhea
in
c h i l d re n
a n d
i m m u n o co m p ro m i s e d
popula5ons.
Infec5ons
are
seasonal
in
nature,
following
the
rainy
season
in
tropical
areas
and
the
winter
season
in
temperate
areas.
The
different
serotypes
have
varying
clinical
presenta5ons,
with
diarrhea,
fever,
and
vomi5ng
being
common
symptoms.
Human
astroviruses
(HAstV)
are
non-‐
enveloped,
single-‐stranded,
posi5ve
sense
RNA
viruses.
The
HAstV
capsid
protein
contains
5
dis5nct
structural
domains.
One
of
these
(known
as
the
acidic
domain),
is
thought
to
be
involved
in
virus
matura5on
and
assembly.
Methods:
• Cloning
was
performed
to
produce
a
pET52b
expression
plasmid
containing
the
acidic
domain
nucleo5de
sequence
• Protein
was
expressed
in
the
BL21
E.
coli
cell
line
• Purifica9on
was
performed
by
affinity
and
size
exclusion
chromatography
• Computa5onal
tools
were
used
to
predict
secondary
and
ter5ary
structure
• Nuclear
magne5c
resonance
(NMR)
and
circular
dichroism
(CD)
were
performed
to
corroborate
predicted
secondary
structural
characteris5cs
• A
trypsin
digest
was
performed
to
isolate
the
puta5ve
helical
bundle
• Screening
trays
were
set
up
to
find
ideal
crystalliza9on
condi5ons
Acknowledgments:
!
! ! Thank you to Professor DuBois for acting as my senior thesis advisor, and to Crown College !
! ! for project funding.
Results:
Cloning,
expression,
and
purifica5on
produced
an
ample
supply
of
stable
acidic
domain
protein.
Conclusions:
While
the
ter5ary
structure
of
the
acidic
domain
is
s5ll
unknown,
protocols
for
successfully
producing
and
purifying
it
have
been
demonstrated
here.
Several
lines
of
evidence
(including
tryp5c
digest
paWern,
circular
dichroism,
and
sizing
behavior)
have
helped
confirm
the
predicted
secondary
structure
of
approximately
40%
helix
and
60%
unstructured.
AWempts
to
crystallize
both
the
en5re
domain
and
the
puta5ve
helices
isolated
by
tryp5c
digest
were
ul5mately
unsuccessful.
!
!
!
Elizabeth
A.
Lagesse
DuBois Lab, University
of
California,
Santa
Cruz
Structural Studies of the Human Astrovirus 1 Capsid Acidic Domain
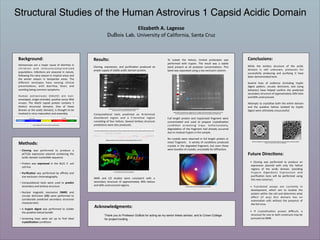
SDS
PAGE
aHer
size
exclusion
chromatography
showing
the
acidic
domain
protein
as
a
bright
band
just
below
25
kDa
Computa5onal
tools
predicted
an
N-‐terminal
disordered
region
and
a
C-‐terminal
region
consis5ng
of
four
helices.
Several
ter5ary
structure
predic5ons
were
also
produced.
RobeLa
secondary
structure
predic9on,
showing
predicted
helices
as
red
“H”s
NMR
and
CD
studies
were
consistent
with
a
secondary
structure
of
approximately
40%
helices
and
60%
unstructured
regions.
2
of
5
RoseLa
ter9ary
structural
predic9ons
To
isolate
the
helices,
limited
proteolysis
was
performed
with
trypsin.
The
result
was
a
stable
band
present
at
all
protease
concentra5ons.
This
band
was
separated
using
a
size
exclusion
column.
Full
length
protein
and
trypsinized
fragment
were
concentrated
and
used
to
prepare
crystalliza5on
condi5on
screening
trays.
Unfortunately,
degrada5on
of
the
fragment
had
already
occurred
due
to
residual
trypsin
in
the
sample.
No
crystals
were
observed
in
full
length
protein
or
intact
fragment.
A
variety
of
condi5ons
produced
crystals
in
the
degraded
fragment,
but
even
these
were
bundles
of
crystals,
unsuitable
for
diffrac5on.
Trypsin
digest
concentra9on
test
ranging
from
10:1
to
2500:1
acidic
domain:trypsin
ra9o
(by
mass).
The
red
arrow
corresponds
to
the
full
length
construct,
and
the
blue
arrow
corresponds
to
the
stable
band
Crystal
bundles
formed
from
degraded
protein
sample
Domain
diagram
for
the
human
astrovirus
capsid
protein
Future
Direc9ons:
!
•
Cloning
was
performed
to
produce
an
expression
plasmid
with
only
the
helical
regions
of
the
acidic
domain.
(avoiding
trypsin
diges5on)
Expression
and
purifica5on
tests
will
be
performed
using
this
new
construct.
!
•
Func5onal
assays
are
currently
in
development,
which
aim
to
localize
the
protein
within
the
cell
and
determine
what
effect
(if
any)
this
domain
has
on
mammalian
cells
without
the
presence
of
the
full
virus.
!
•
If
crystalliza5on
proves
difficult,
a
structure
for
one
or
both
constructs
may
be
pursued
via
NMR.
Electron
micrographs
of
a
single
immature
astrovirus
(leH)
and
a
group
of
mature
astroviruses
(right).
(Dreyden
et.
al.
2012,
Crea9ve
Commons
2.0,
respec9vely)