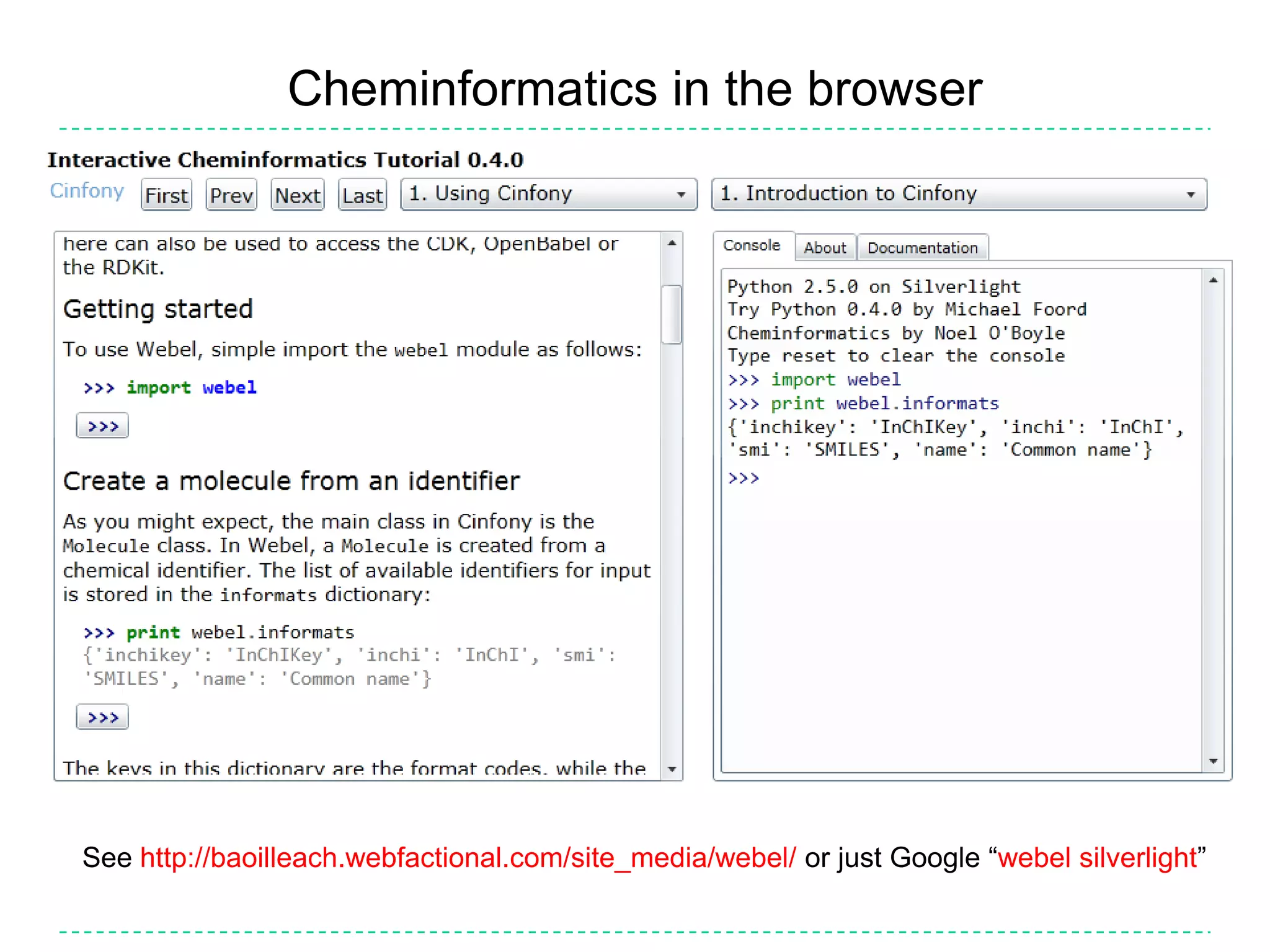
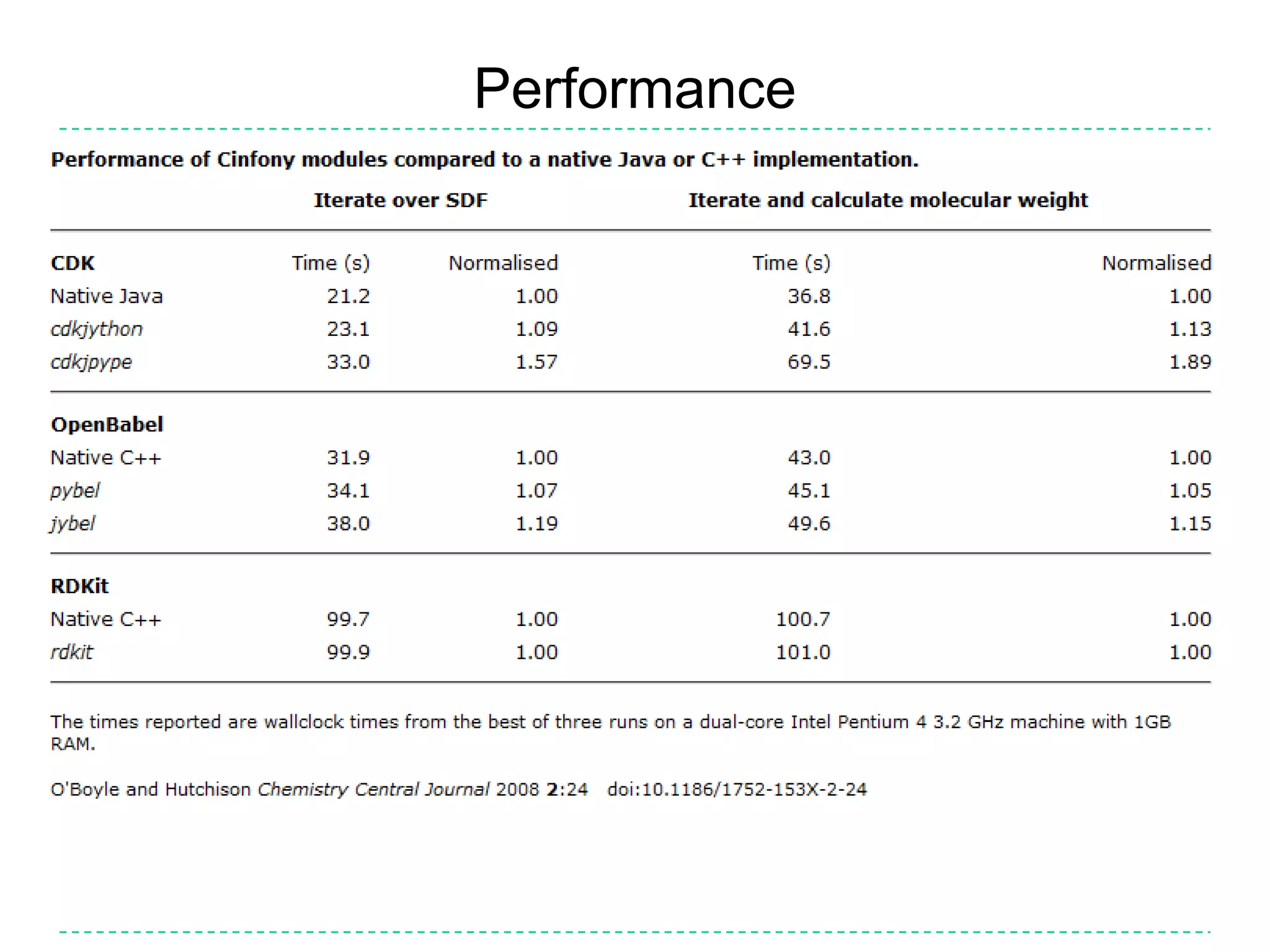
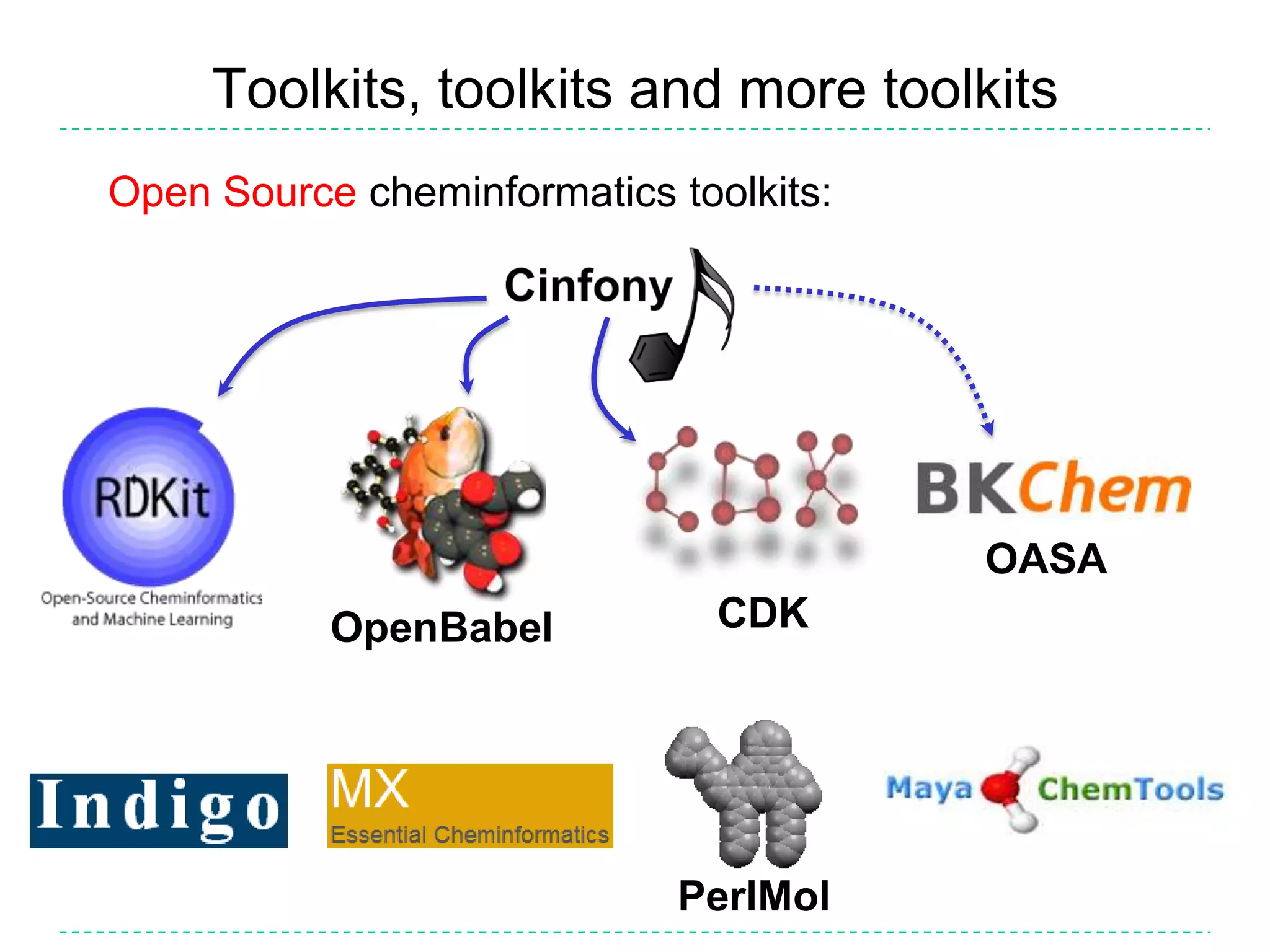
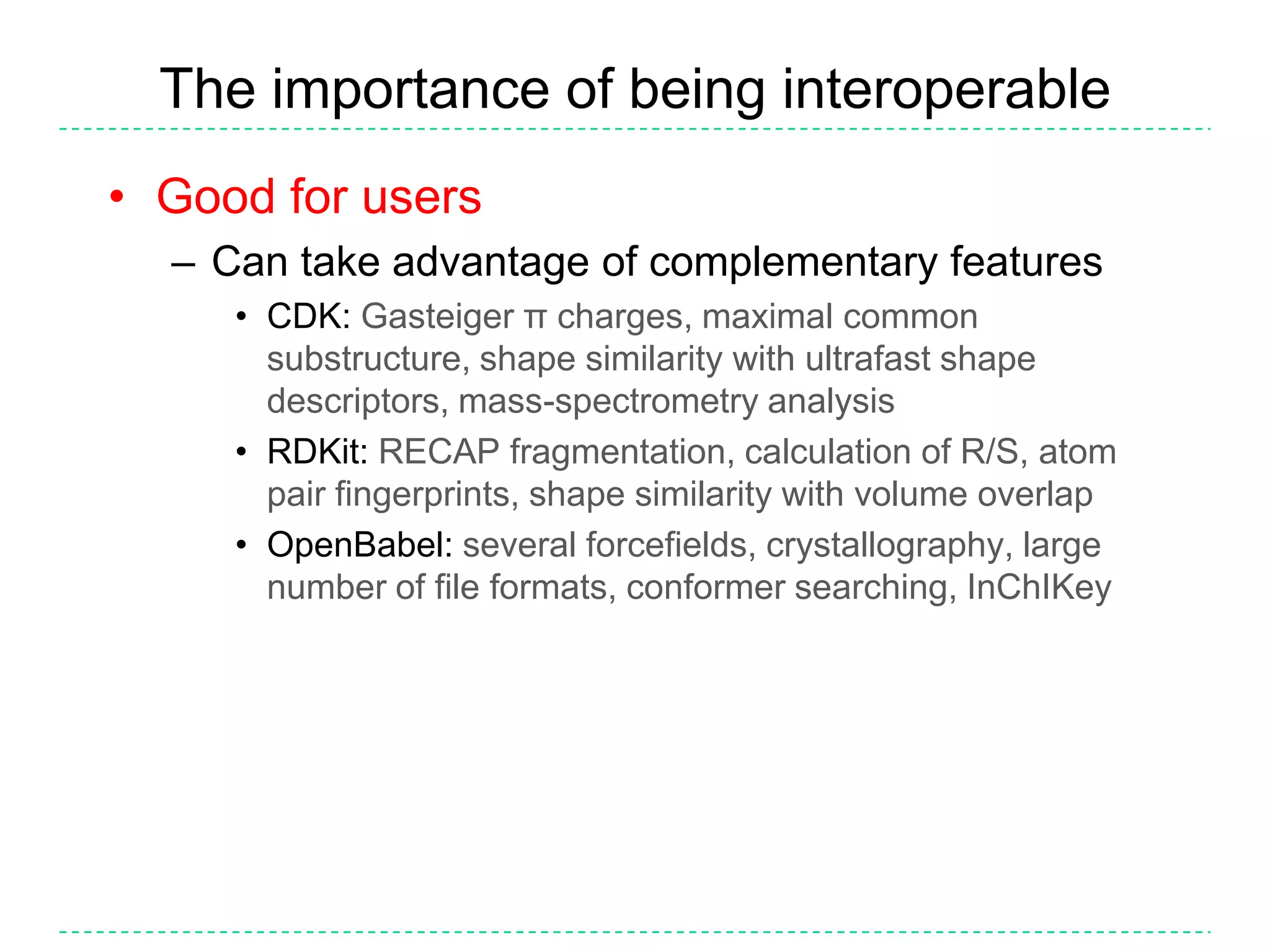
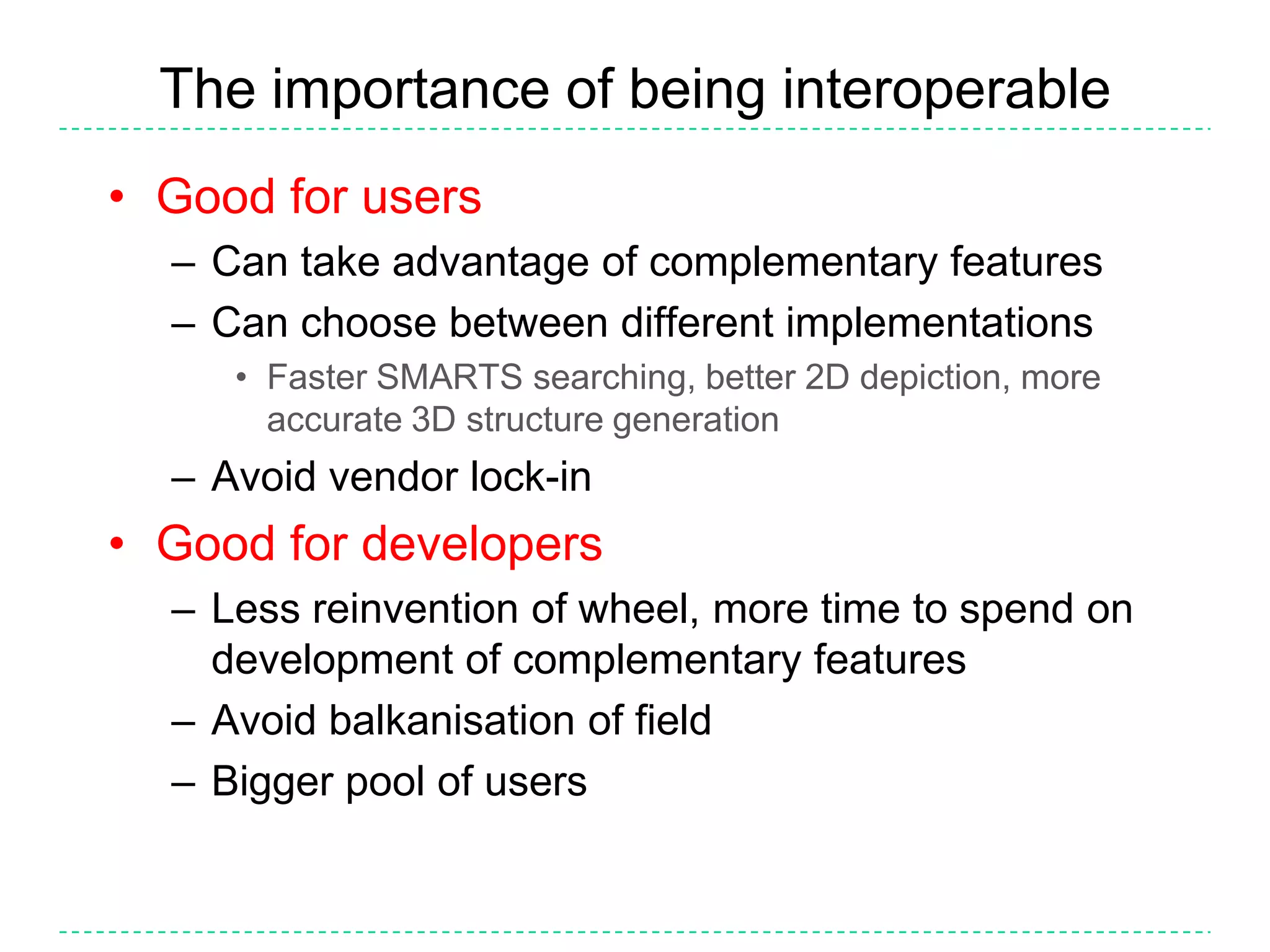
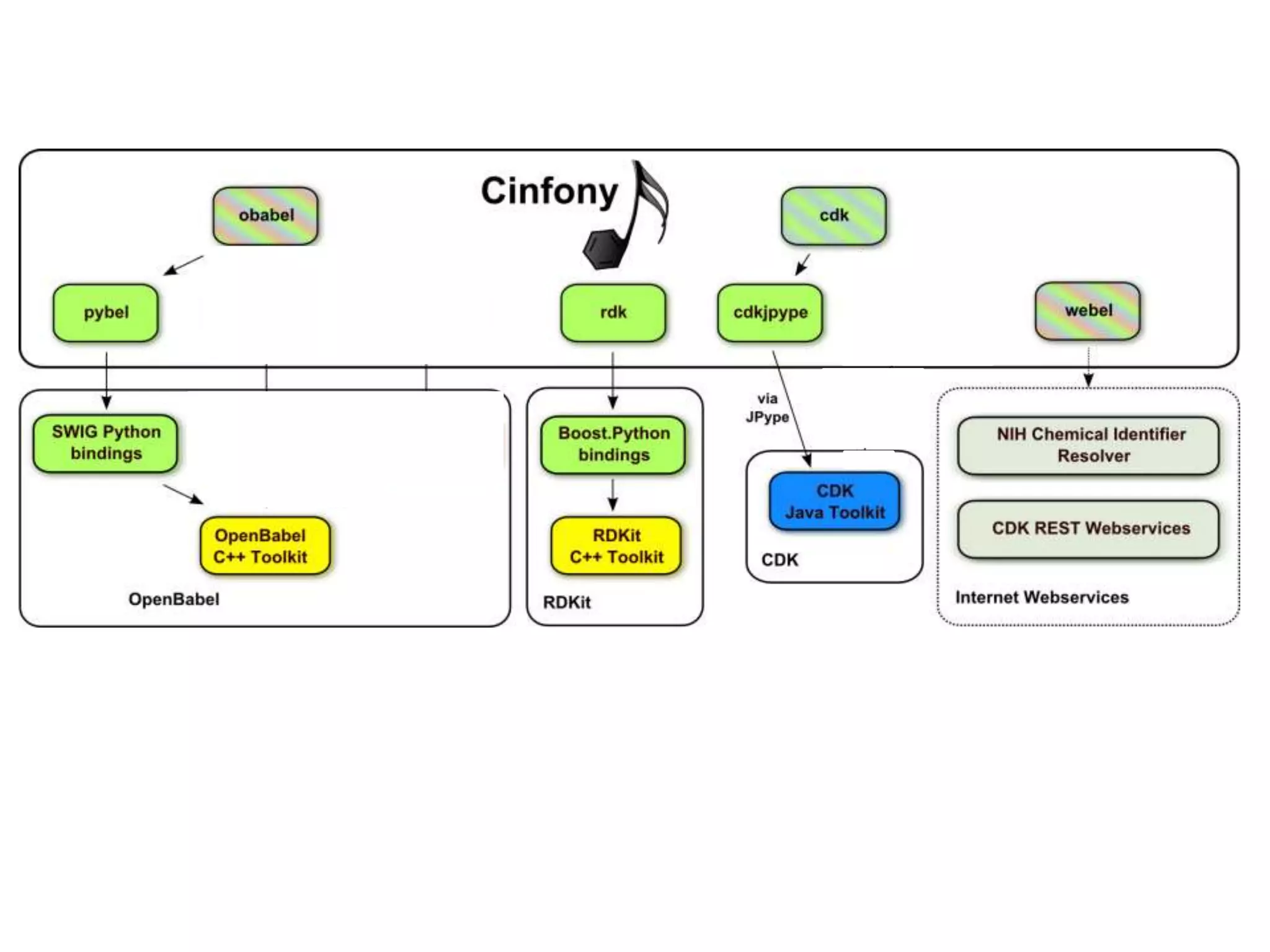
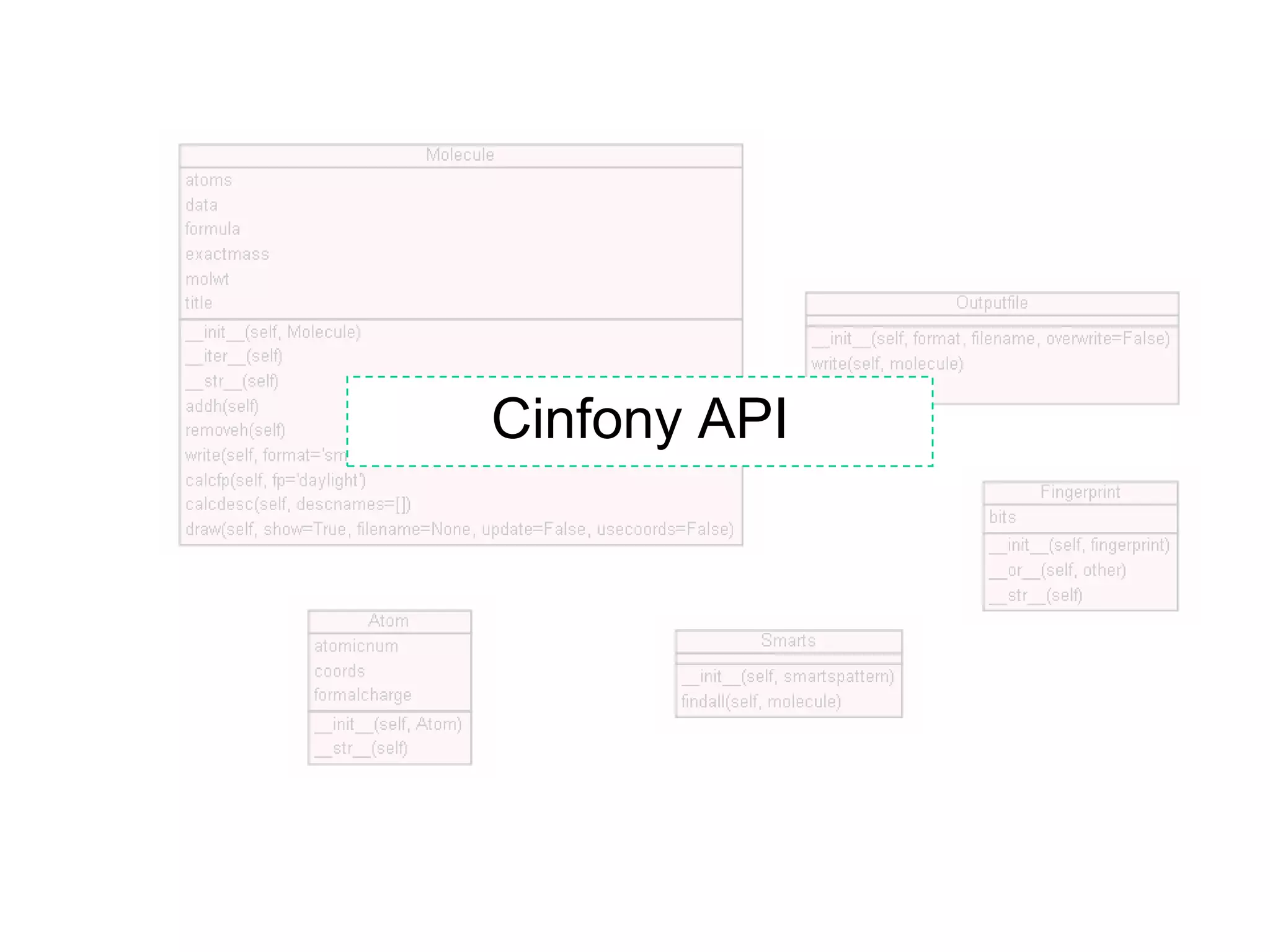
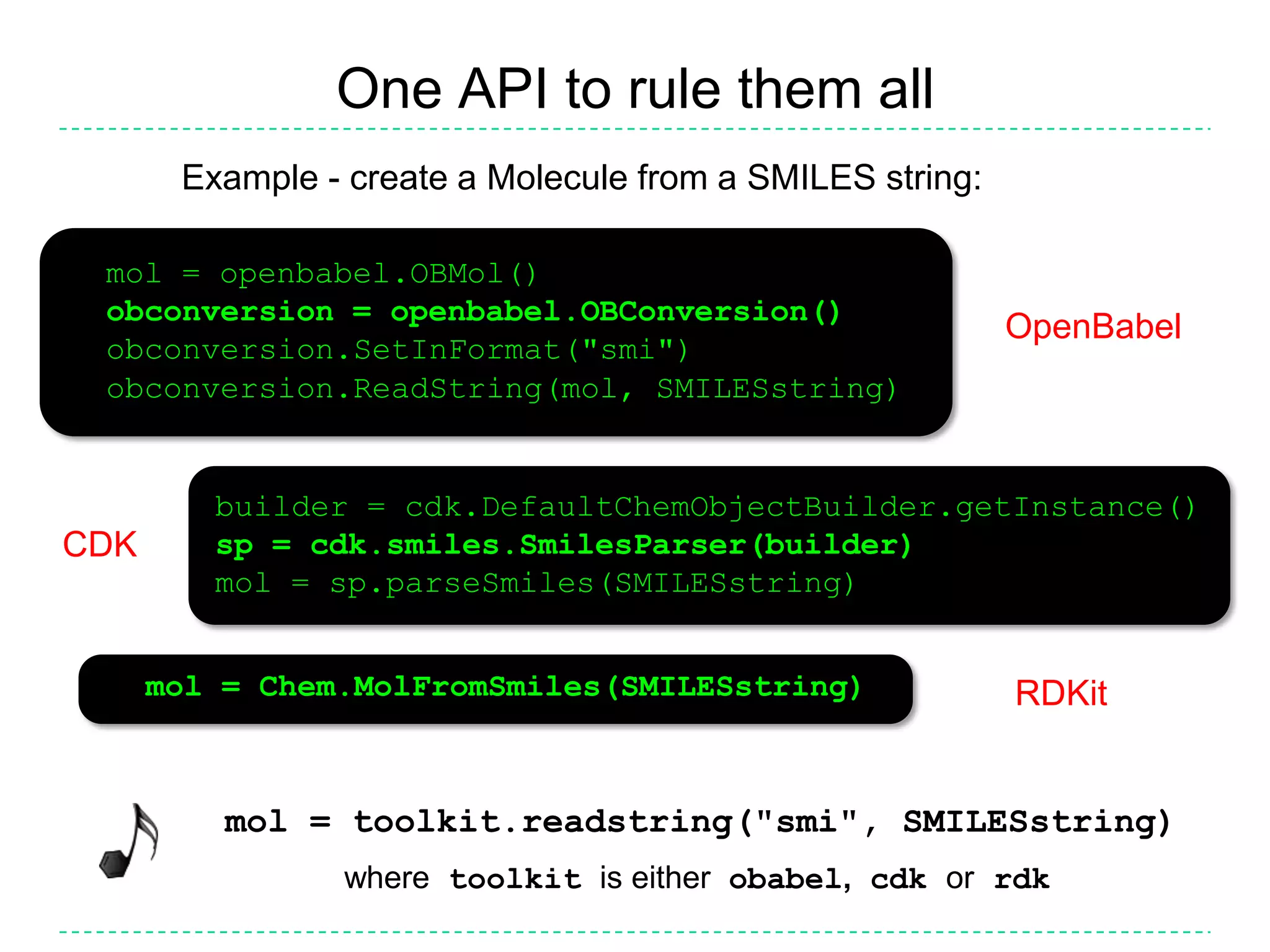
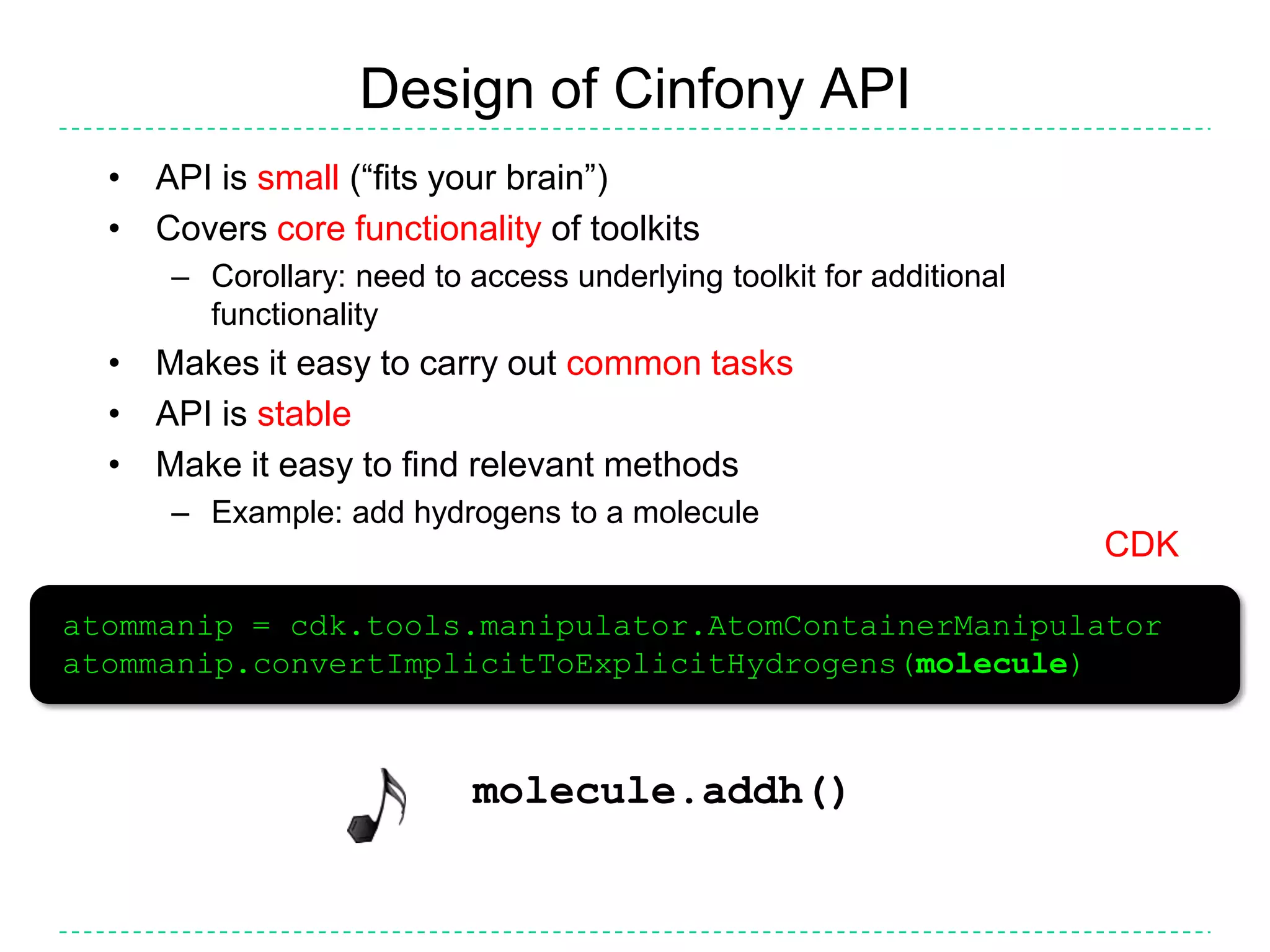
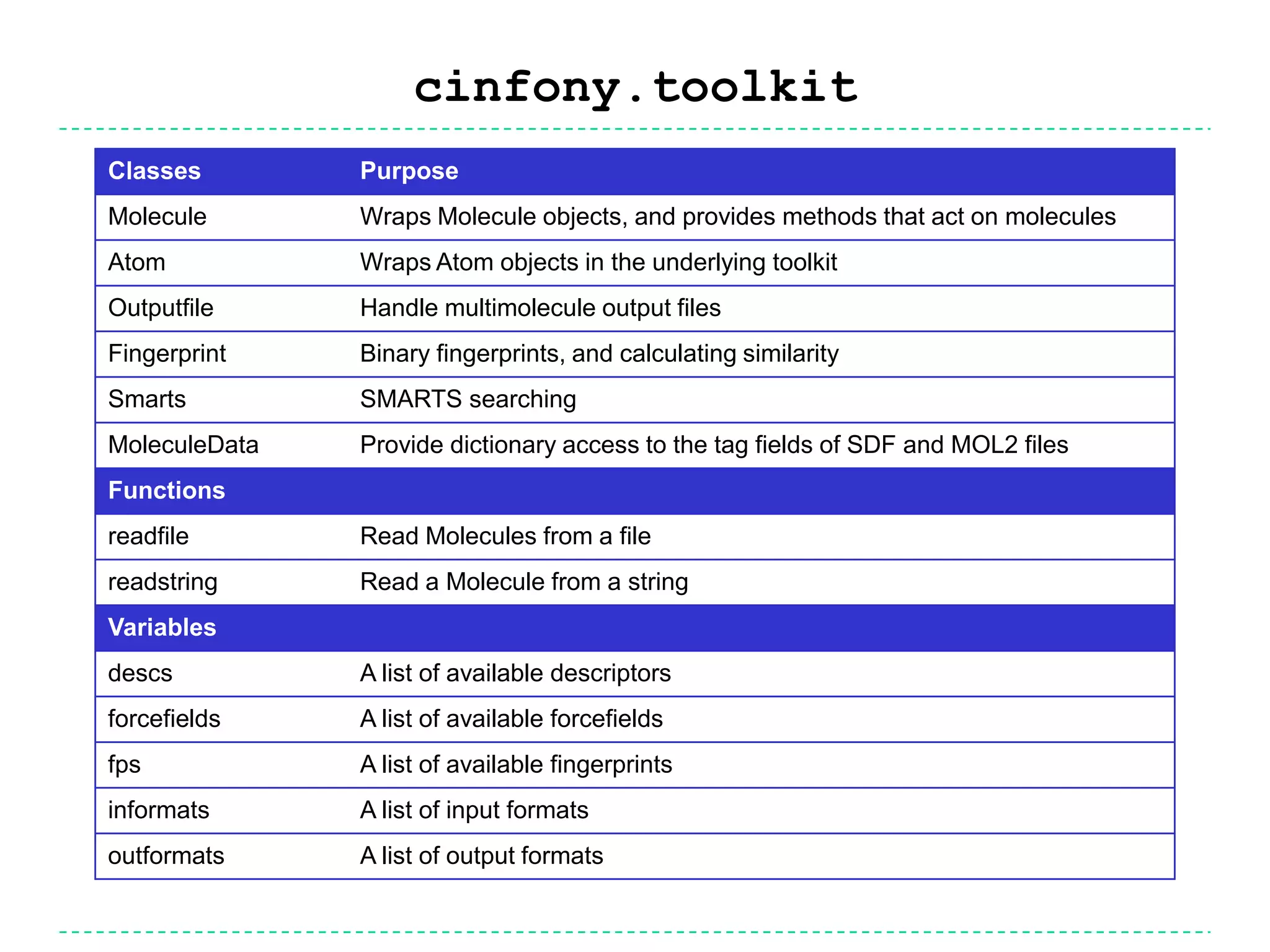
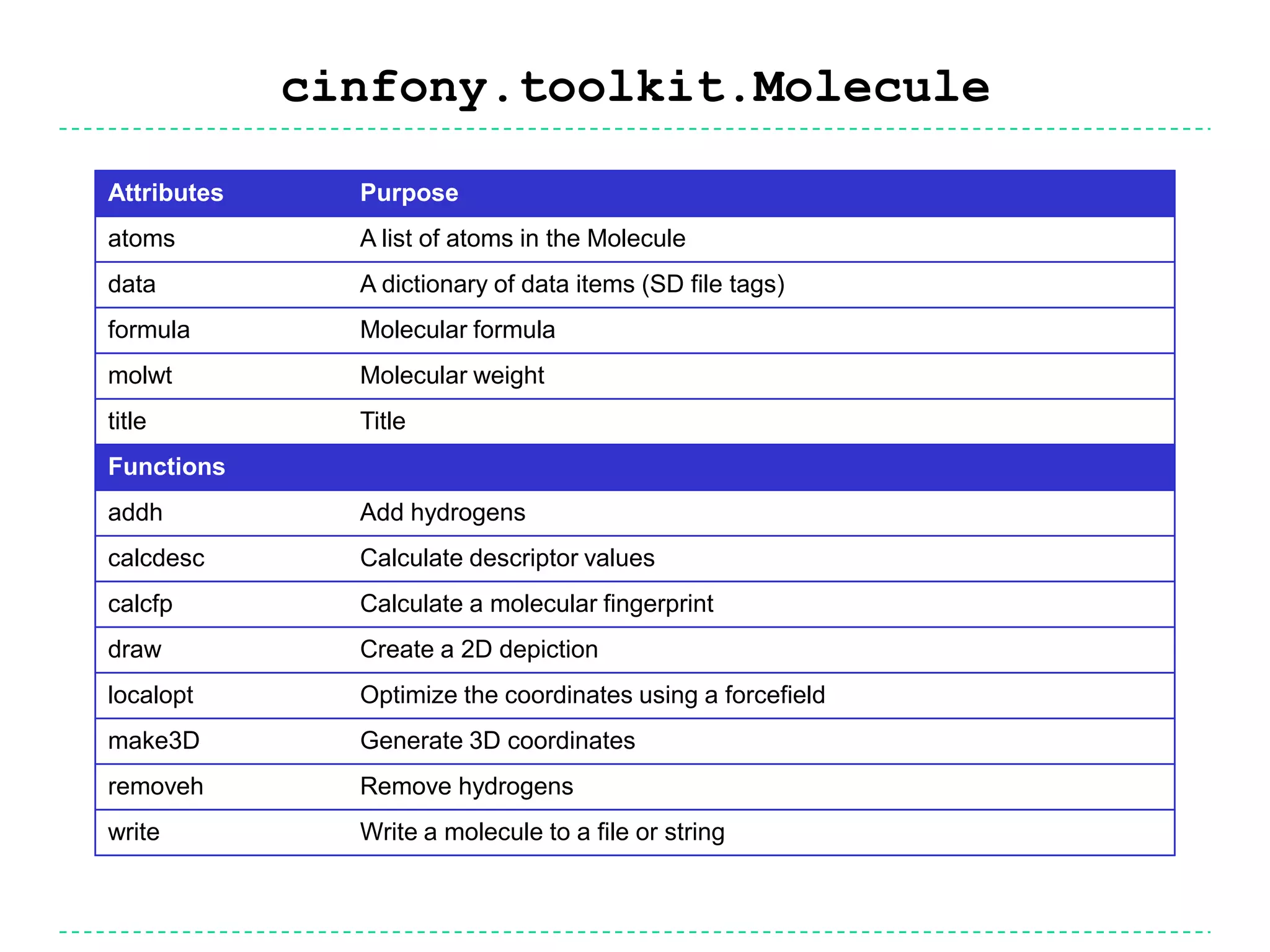
The document discusses the integration of various cheminformatics toolkits into a unified framework called Cinfony to enable interoperability and enhance user experience. It highlights the strengths of multiple toolkits such as CDK, Open Babel, and RDKit, emphasizing the benefits of using a common API for seamless functionality. The text also introduces Webel, a Cinfony module that operates via web services, facilitating cheminformatics tasks directly through a web browser.




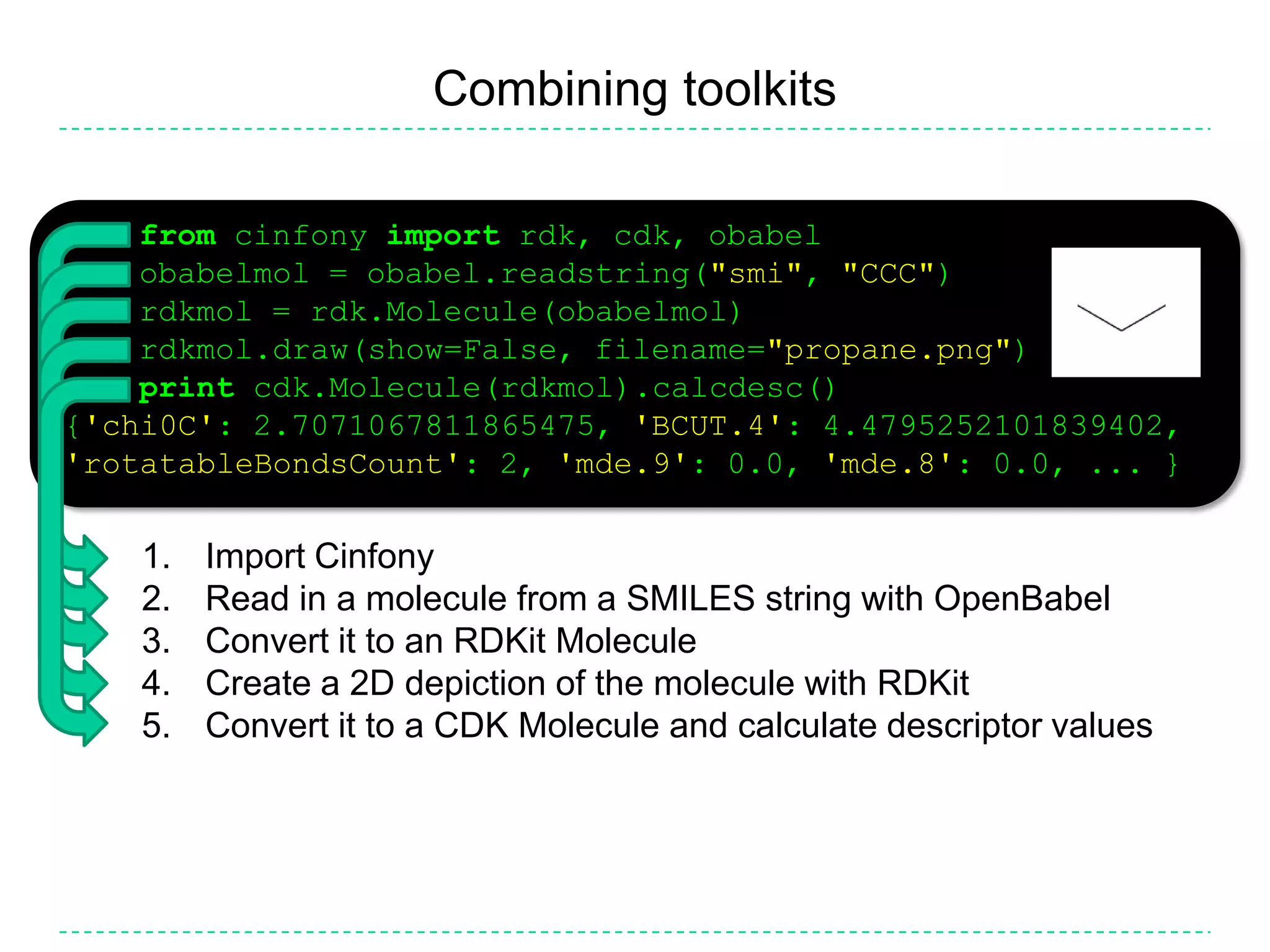

![Comparing toolkits>>> from cinfony import rdk, cdk, obabel>>> for toolkit in [rdk, cdk, obabel]:... mol = toolkit.readstring("smi", "CCC")... print mol.molwt... mol.draw(filename="%s.png" % toolkit.__name__)Import CinfonyFor each toolkit...... Read in a molecule from a SMILES string... Print its molecular weight... Create a 2D depictionUseful for sanity checks, identifying limitations, bugsCalculating the molecular weight (http://tinyurl.com/chemacs3)implicit hydrogen, isotopesComparison of descriptor values (http://tinyurl.com/chemacs2)Should be highly correlatedComparison of depictions (http://tinyurl.com/chemacs1)](https://image.slidesharecdn.com/cinfonyacs-100325191015-phpapp02/75/Cinfony-Combining-disparate-cheminformatics-resources-into-a-single-toolkit-18-2048.jpg)