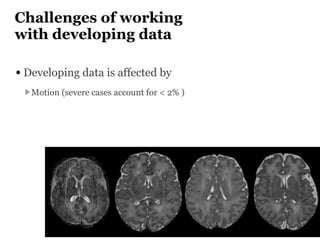
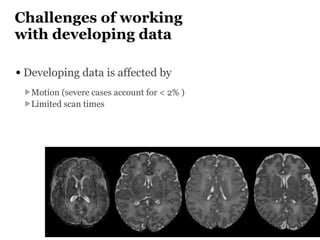
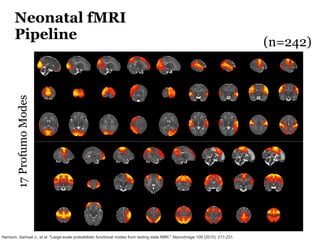
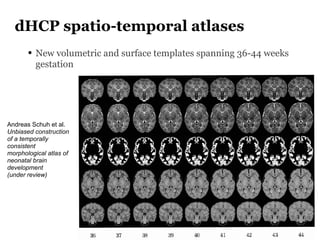
The Developing Human Connectome Project (DHCP) aims to map brain connectivity from 20-44 weeks post-menstrual age using approximately 1500 MRI scans and additional genetic and cognitive data. The project faces challenges such as motion artifacts, limited scan times, and relatively low resolution while employing advanced techniques in motion correction and surface matching. Additionally, it is developing spatio-temporal atlases and utilizing deep learning methods for motion artifact detection in neonatal diffusion MRI.