More Related Content
Similar to CRISPRposter2015Biol446
Similar to CRISPRposter2015Biol446 (20)
CRISPRposter2015Biol446
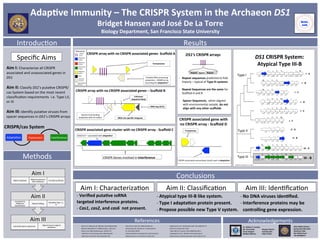
- 1. Adap%ve(Immunity(–(The(CRISPR(System(in(the(Archaeon(DS1((
Bridget(Hansen(and(José(De(La(Torre(
Biology(Department,(San(Francisco(State(University((
Methods(
Introduc.on(
References(
Koonin(EV,(Makarova(KS.(RNA(Biol.(2013;10(5):679K86.(
Bikard(D,(Marraffini(LA.(F1000Prime(Rep.((2013;5:47.(
Grissa(I(et(al.(BMC(Bioinforma.cs.(2007;8:172.(
(HaX(DH(et(al.(PLoS(Comput(Biol.(2005;1(6):e60.(
(Hale(CR(et(al.(Mol(Cell.(2012;45(3):292K302.(
((
Hale(CR(et(al.(Mol(Cell.(2009;139:945–56.(
Hochstrasser(ML,(Doudna(JA.(Trends(Biochem((
Sci.(2015;40(1):58K66.(
Jiang(W,(Bikard(D.(Nat(Biotechnol.(2013;31:233–9.(
Makarova(KS(et(al.(Front(Genet.(2014;5:102.(
(
Makarova(Ks(et(al.((Nat(Rev(Microbiol.(2011;9(6):467K77.(
Rath(D(et(al.(Biochimie.(2015(
Taylor(DW(et(al.(Science.(2015;348(6234):581K5.(
Vestergaard(G(et(al.((RNA(Biol.(2014;11(2):156K67.(
Westra(ER(et(al.(Nat(Rev(Microbiol.(2014;12(5):317K26.(
((
Results(
Acknowledgements(
Nature Reviews | Microbiolo
cas1cas9 cas2 cas4
cas1 cas2cas9 csn2
cas5cas7cas8b cas1cas4cas6 cas2
cas5cas3 cas1cas4cas7cas8c cas2
cas5cas7cas8a1 cas3′cas3′′
cas3′cas3′′
cas8a2csa5cas1 cas4 cas6cas2
csy1 csy2 csy3 cas6fcas2–cas3cas1
cse1 cse2 cas7 cas5cas3 cas1 cas2cas6e
cas10 cmr3 cmr4 cmr5 cmr6cas6cmr1
cas10d csc2 csc1 cas6cas3 cas1cas4 cas2
cas1csm6csm5csm4csm3csm2cas10cas6 cas2
cas1 cas2
L R RRE
L RRE
L LS R R RE
I-A (ApernorCASS5)
L R R RE
L S R R RE
REL SR R RR
L RER R
RE L S R R R T
II-B (NmeniorCASS4a)
II-A (NmeniorCASS4)
I-B (Tneap–HmariorCASS7)
I-C (DvulgorCASS1)
I-F (YpestorCASS3)
I-E (EcoliorCASS2)
III-B (Polymerase–RAMPmodu
I-D
III-A (MtubeorCASS6)
signature genes (TABLE 2). However, for
the loci that cannot be classified even at
the type level, such as the CRISPR–Cas
system in Acidithiobacillus ferrooxidans
str. ATCC 23270 (discussed further below),
we propose the name type U.
Distribution of the three types of CRISPR–
Cas systems in the Archaea and the Bacteria.
The three types of CRISPR systems show a
distinctly non-uniform distribution among
the major lineages of the Archaea and the
Bacteria (TABLE 1). In particular, the type II
systems have been found exclusively in the
Bacteria so far, whereas type III systems are
more common in the Archaea. The previ-
ously observed trend of over-representation
of CRISPR in the Archaea compared to the
Bacteria still holds14,39
(TABLE 1). Moreover,
!"#$%&'(')!"#$!%$&'()*+,#)-!*.!(#$!(#%$$!/'0*%!(1-$,!'+2!($+!,345
(1-$,!*.!6789:7!,1,($/,;!*+&',-."/012'3"4.1&3,'5.&%56'0%/+",&/,$%&3'
0%&'3+576'85%'&0/+',-.&'069'3$:,-.&'58';<=>?<@;03'A/1$3,&%&9'%&#$10%1-'
"6,&%3.0/&9'3+5%,'.01"69%54"/'%&.&0,3@;<=>?<B0335/"0,&9'.%5,&"63C'
3-3,&4D'6$4&%5$3'E0%"0,"563'&F"3,G'H%,+515#5$3'#&6&3'0%&'/515$%'/59&9'
069'"9&6,"8"&9':-'0'804"1-'604&2'03'#"E&6'"6'TABLE 2G'*+&'3"#60,$%&'#&6&3'
85%';<=>?<@;03',-.&3'0%&'3+576'7",+"6'#%&&6':5F&32'069',+53&'85%'3$:B
,-.&3'0%&'3+576'7",+"6'%&9':5F&3G'*+&'1&,,&%3'0:5E&',+&'#&6&3'3+57'
40I5%'/0,%"&3'58';03'.%5,&"63J'10%#&';<=>?<B0335/"0,&9'/54.1&F'85%'
06,"E"%01'9&8&6/&'A;03/09&C'3$:$6",3'AKC2'34011';03/09&'3$:$6",3'A>C2'
%&.&0,B0335/"0,&9'4-3,&%"5$3'.%5,&"6'A<LM?C';03/09&'3$:$6",3'A<C2'
<LM?'804"1-'<N03&3'"6E51E&9'"6'/%<NL'.%5/&33"6#'A<OC'A65,&',+0,'561-'
069',%063/%".,"5601'%&#$10,5%3'A*CG'*+&'3,0%'"69"/0,&3'0'.%&9"/,&9'"60
!"#$"%'069'!"#$"&'#&6&3'0%&',-."/011-'4$,$011-'&F/1$3"E&':$,':5,+'/
,+&'!"#%'069'!"#&'#&6&3'"6'903+&9':5F&3'0%&'65,'0335/"0,&9'7",+'
90,02',+"3'3/+&40,"/'3+573';03P'A;HQRSTPC'03'0'4&4:&%'58',+&'<LM
3$.&%804"1-G'!5%'&0/+';<=>?<@;03'3$:,-.&'A&F/&.,'85%',+&'6&71-'"9&6
8"&9'3$:,-.&'=BUC2',+&'519'604&3'8%54'REFS 13,14'0%&'"69"/0,&9'"6'.0%&
,+&3&3G'!"#$%&'"3'459"8"&92'7",+'.&%4"33"562'8%54'REF. 14'©'A(VV
!"#$!"%&'("
NATURE REVIEWS | <867=>8=?=@A! VOLUME 9 | JUNE 2011 | !
© 2011 Macmillan Publishers Limited. All rights reserved
I(F(A(
I(F(B(
I(F(C(
I(F(D(
I(F(E(
I(F(F(
II(F(A(
II(F(B(
III(F(A(
III(F(B(
COG1517
COG1517
Dr.(William(P.(Cochlan(
Mr.(Chris(Ikeda((
Chuck(Wingert(
Maribel(Albarran(
(
(
(
Morgan(Meyers(
Kate(BarreNo(
Alison(Fisher(
(
2015(MARC(Scholars(
Spring(2015(Microbial(
Genomics(Class(
NIH(MARC(Grant(
T34FGM008574(
Conclusions(((
K Verified(puta%ve(ssRNA(
targeted(interference(proteins.(
K Cas1,(cas2,(and(cas6((not(present.(
*
(
K Atypical(type(IIIFB(like(system.((
K Type(I(adapta%on(protein(present.(
K Propose(possible(new(Type(V(system.(((
(
(
(
K No(DNA(viruses(iden%fied.(
K Interference(proteins(may(be(
controlling(gene(expression.(
Aim(I:(Characteriza.on( Aim(II:(Classifica.on( Aim(III:(Iden.fica.on(
CRISPR(array(with(no(CRISPR(associated(genesF(Scaffold(A(
CRISPR(array(with(no(CRISPR(associated(genes(–(Scaffold(B(
(
K Repeat(sequences(predicted(to(fold(
linearly(–(typical(of(Type(III(systems(
(
K Repeat(Sequences(are(the(same(for(
Scaffold(A(and(B((
(
K Spacer(Sequences,((when(aligned(
with(environmental(sample,(do(not(
align(with(any(other(scaffolds(
||||
Puta.ve(RNA(processing(
proper.es(–(CRISPR(array(
recrui.ng(for(adapta%on?((
tRNA(site(specific(Integrase(
tRNA(Asp((GTC)(
||||
Unknown(
Sequence(Read(
CRISPR(associated(gene(cluster(with(no(CRISPR(array(F(Scaffold(C((
CRISPR(Genes(involved(in(Interference((
CRISPR(associated(gene(with(
no(CRISPR(array(F(Scaffold(D(
Transposase(
COG1517(–(associated(with(adapta%on((
Repeat(((Spacer((((Repeat(
||||Transposases(
DS1’s(CRISPR(arrays(
Nucleic(Acid(binding(
proper.es(with(Zn(ribbon((
CRISPR(associated(exonuclease((cas4)(used(in(adapta%on(
DS1*CRISPR(System:((
Atypical(Type(IIIFB(
Type(I(
Type(II(
Type(III(
Aim(III(
Extracted(spacer(sequences(
Aligned(spacers(against(
databases(
Aim(II(
Compared(to(
Makarova+
Classifica.on(
Repeat(Folding(
Classified(Type(I,(II,(
III(
Aim(I(
IMG/m(database((
Aligned(sequences(
with(databases(
Func.on(predicted(
Aim(I:(Characterize(all(CRISPR(
associated(and(unassociated(genes(in(
DS1+
+
Aim(II:(Classify+DS1’s(puta.ve(CRISPR/
cas(System(based(on(the(most(recent(
classifica.on(requirements((i.e.(Type(I,II,(
or(III((
(
Aim(III:(Iden.fy(puta.ve(viruses(from(
spacer(sequences(in(DS1’s(CRISPR(arrays((
(
Specific(Aims(
CRISPR/cas(System(
Adapta.on( Expression( Interference(
|||| CRISPR(
array(
Adapta.on(
Protein(
Expression(
Protein(
Interference(
Protein(
Integra.on(
Protein(
tRNA(
Unknown(
Func.on(
Hypothe.cal(
Protein(
Elements(
Adapted(from(Makarova(et(al(2015(