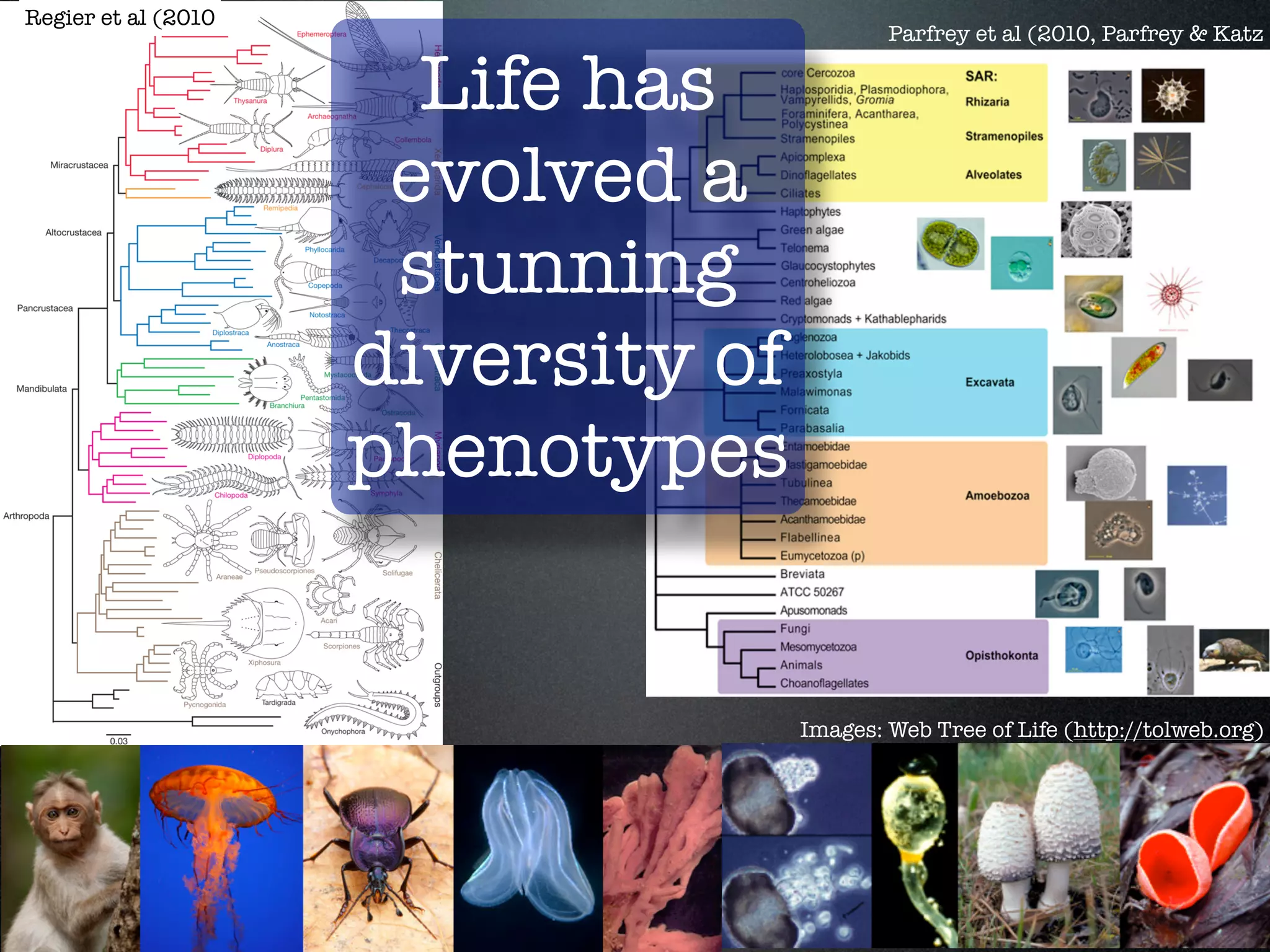
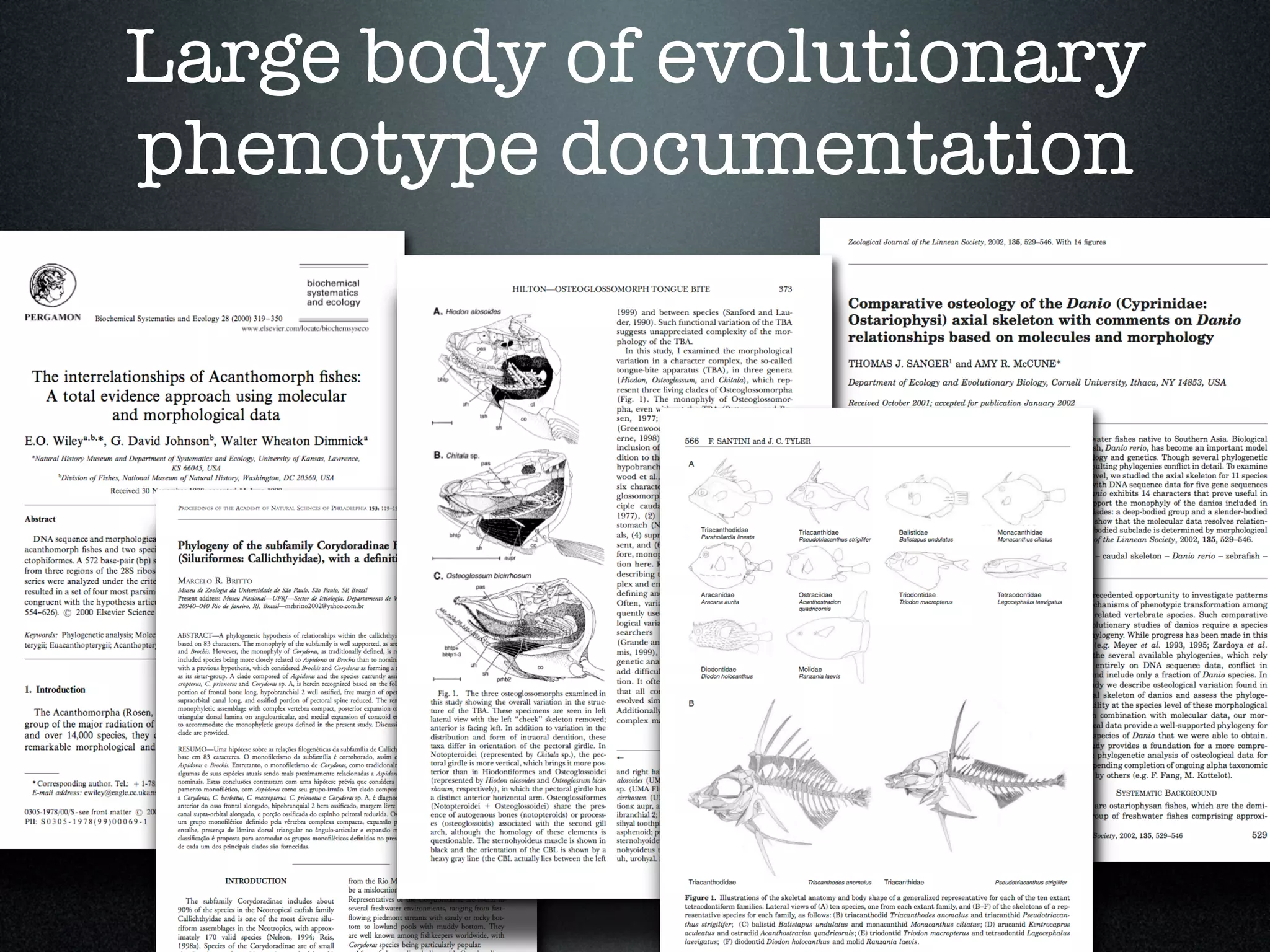
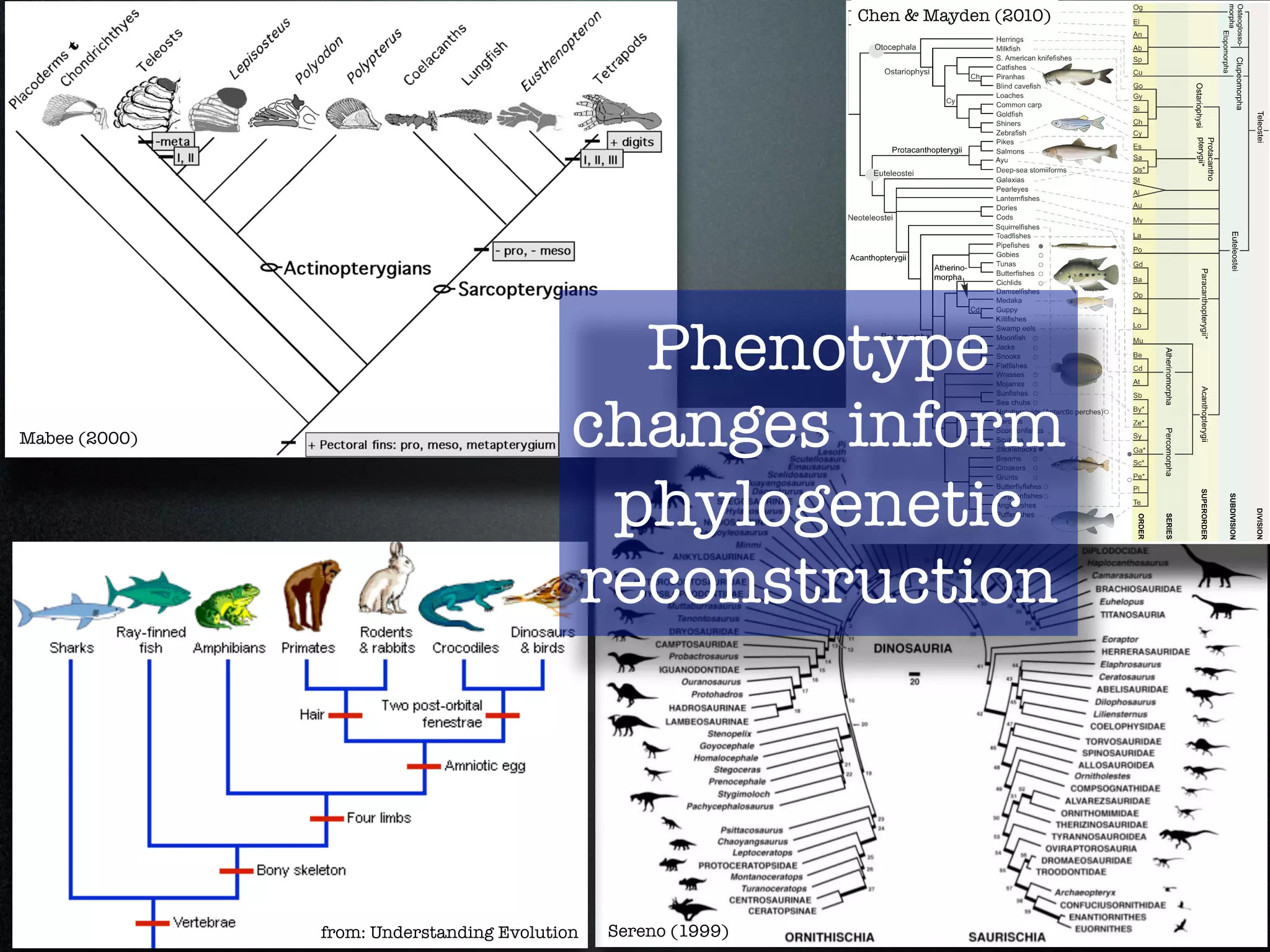
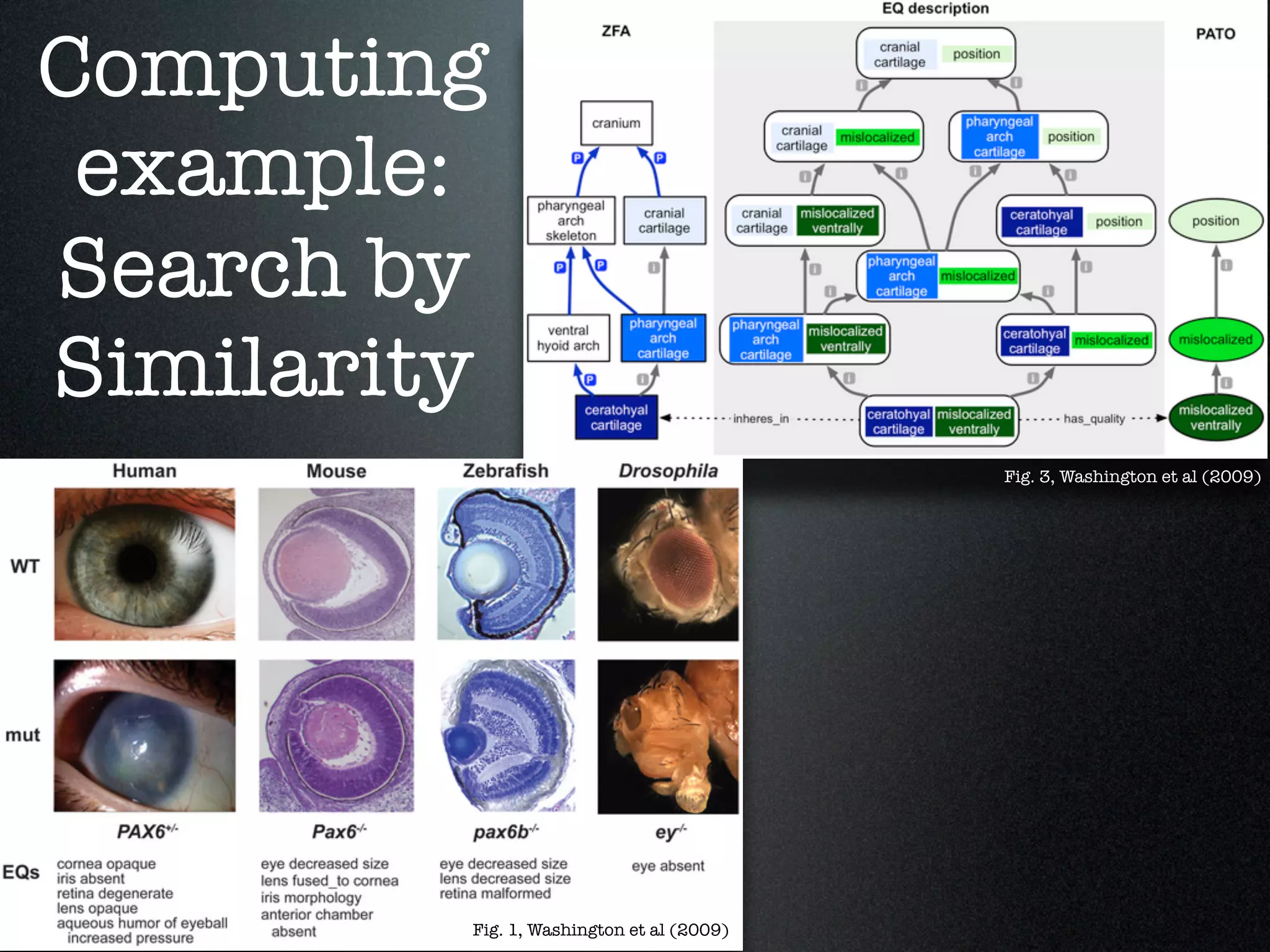
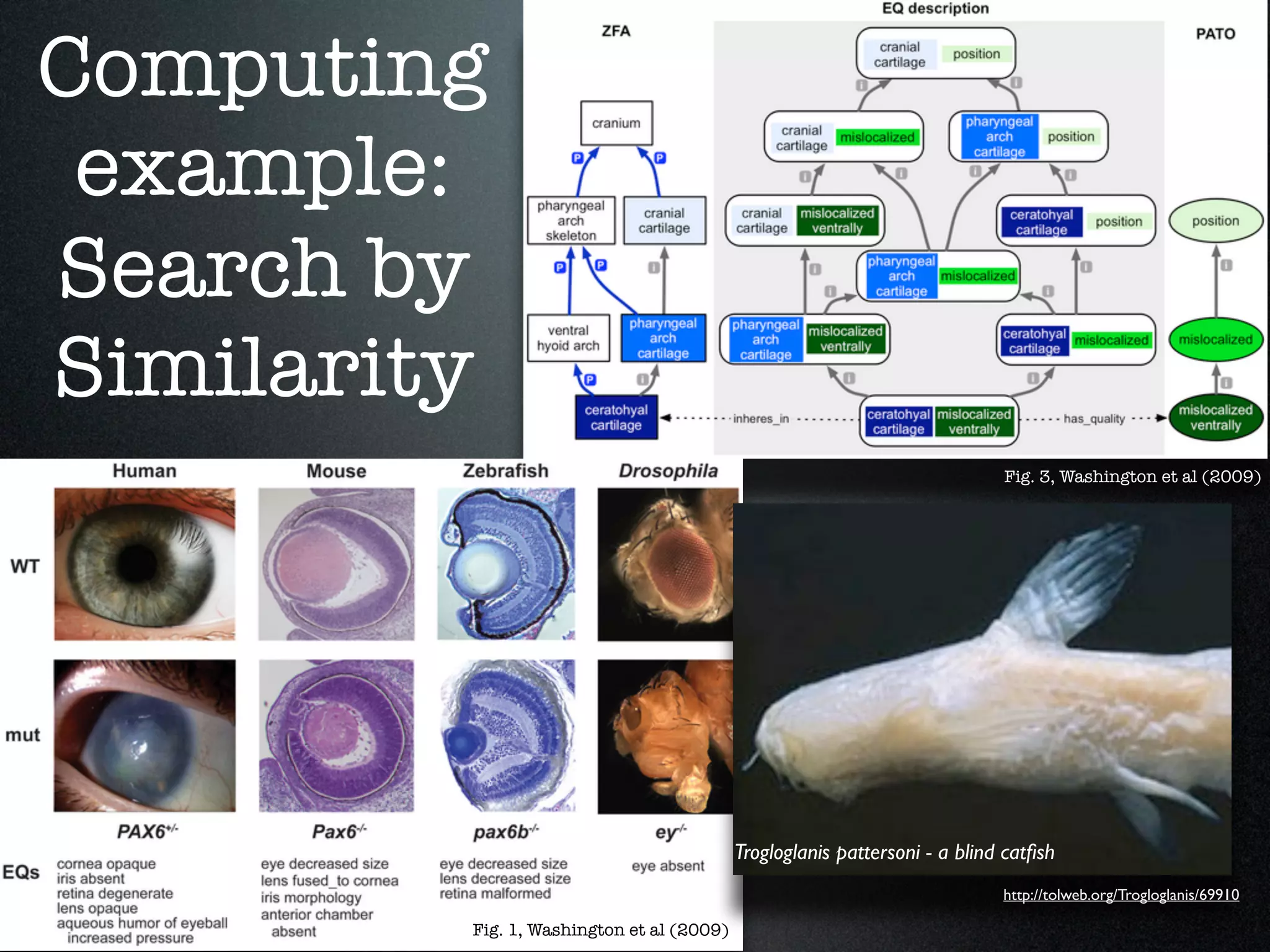
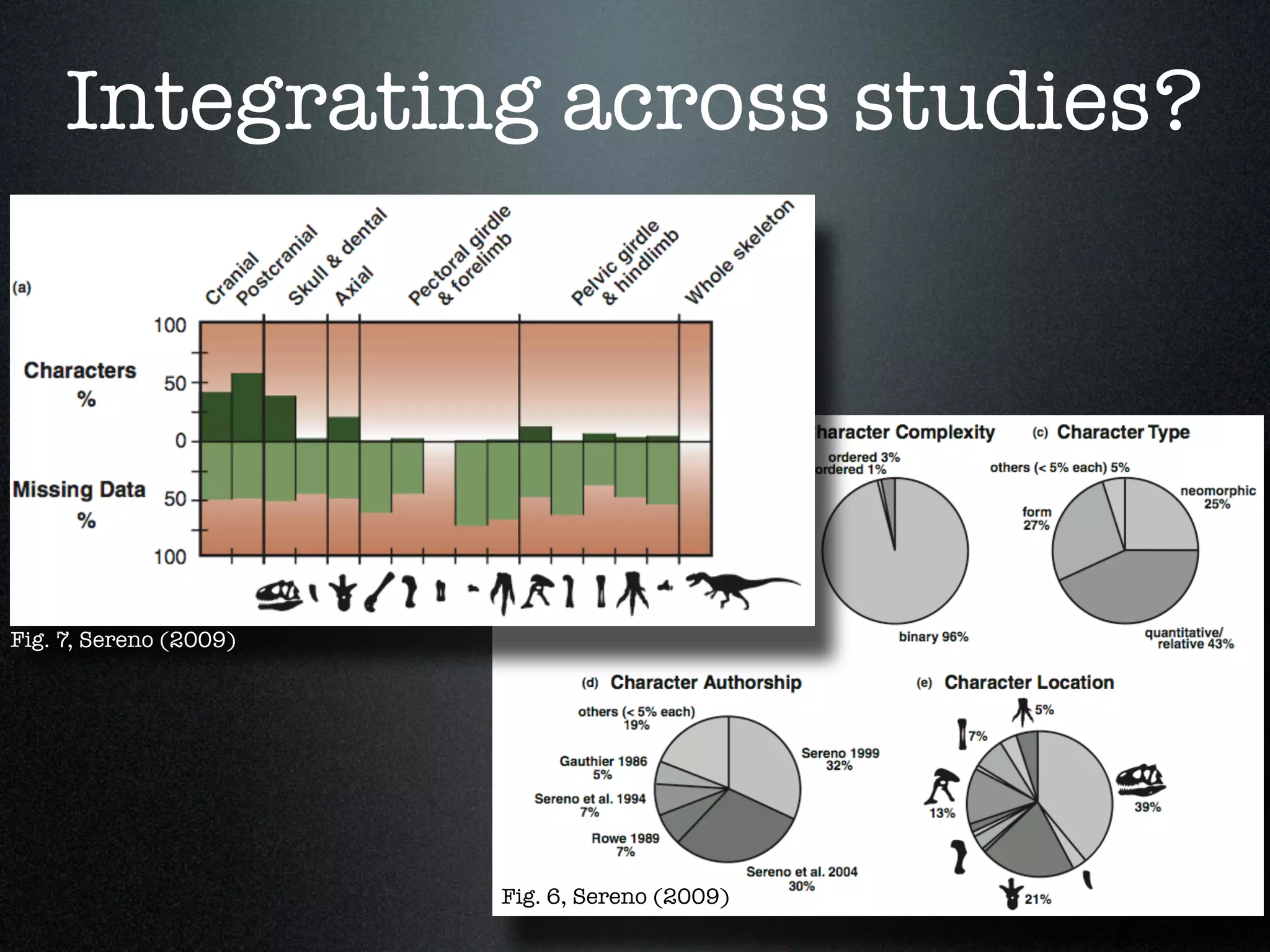
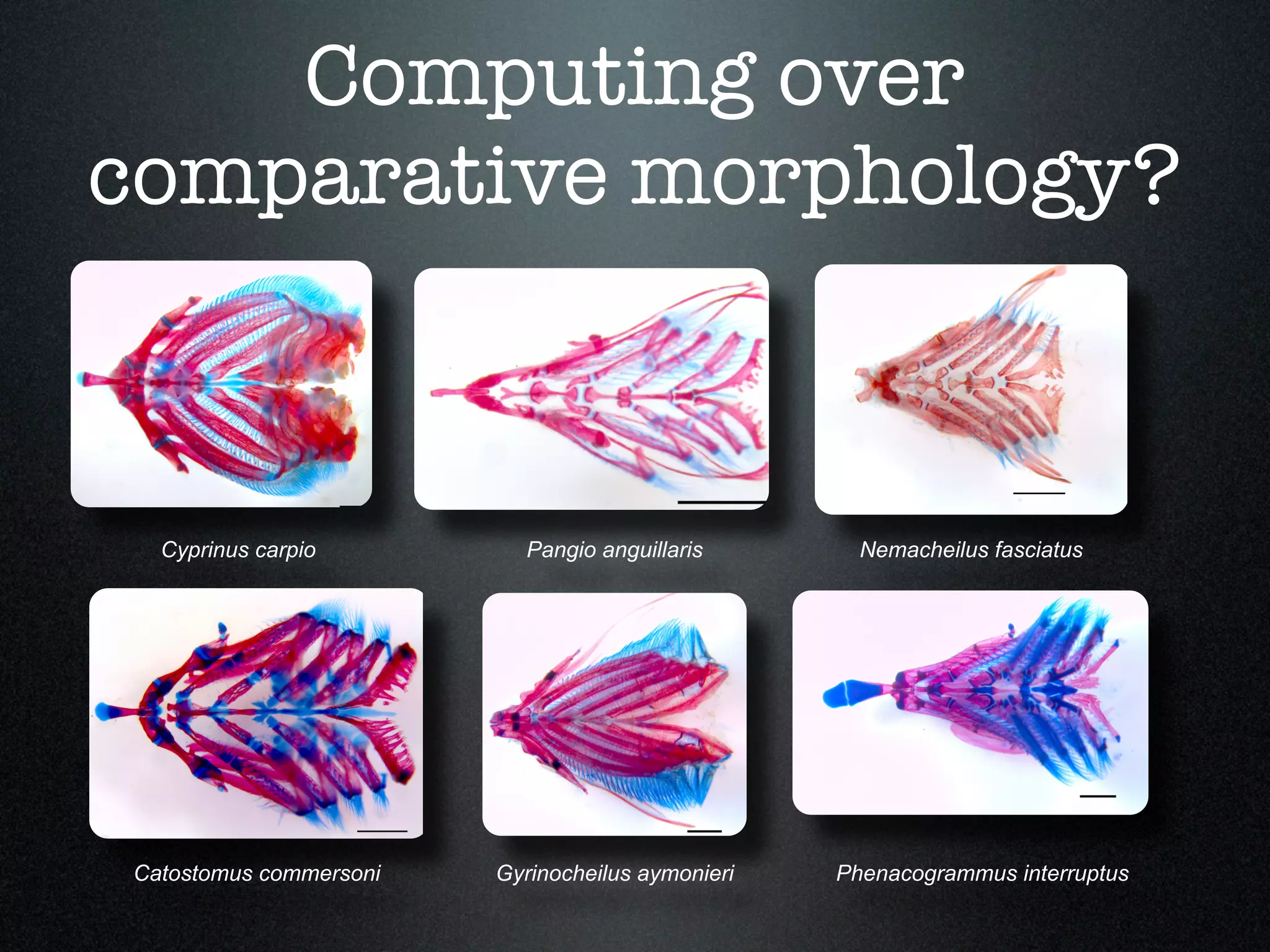
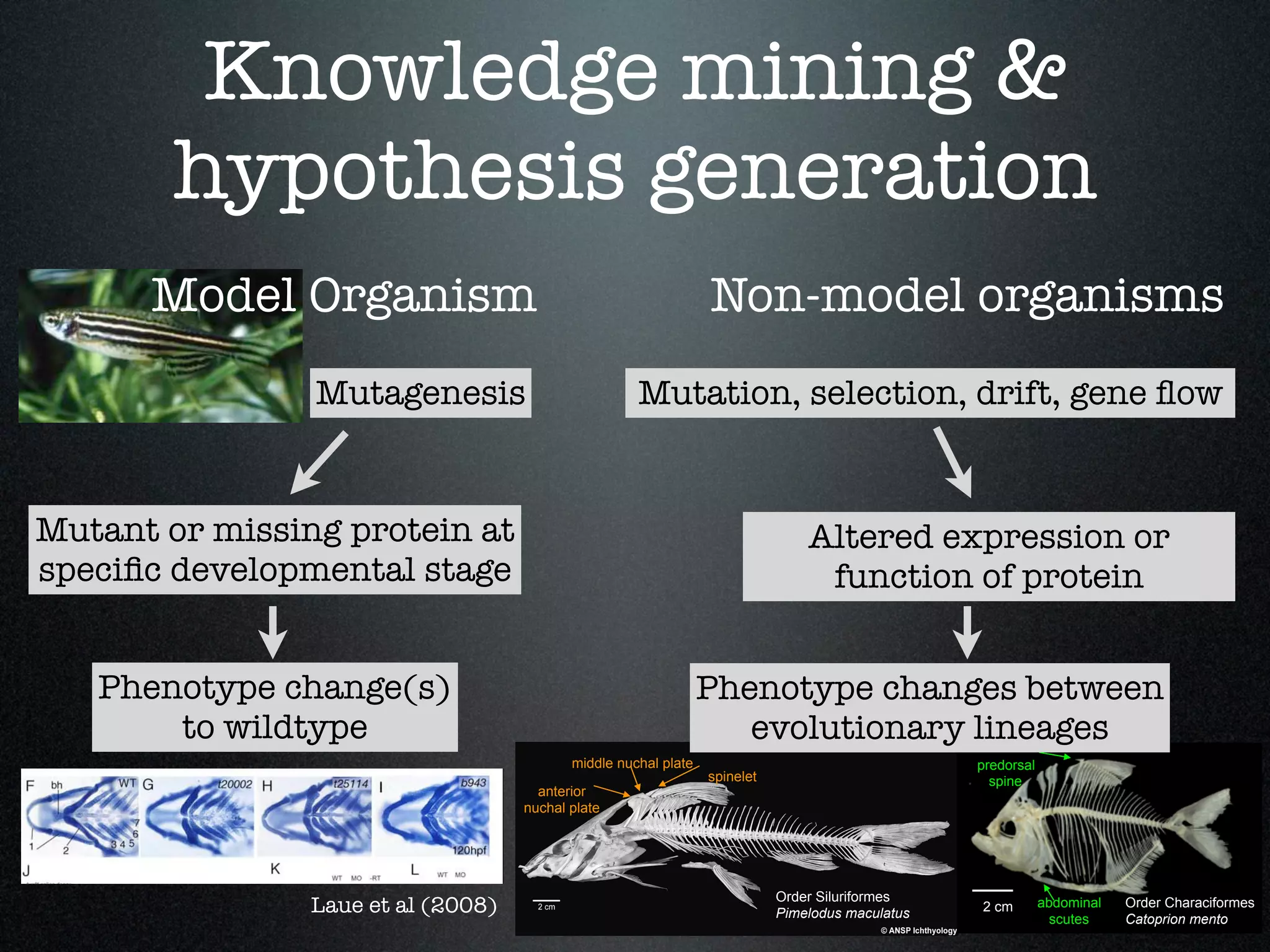
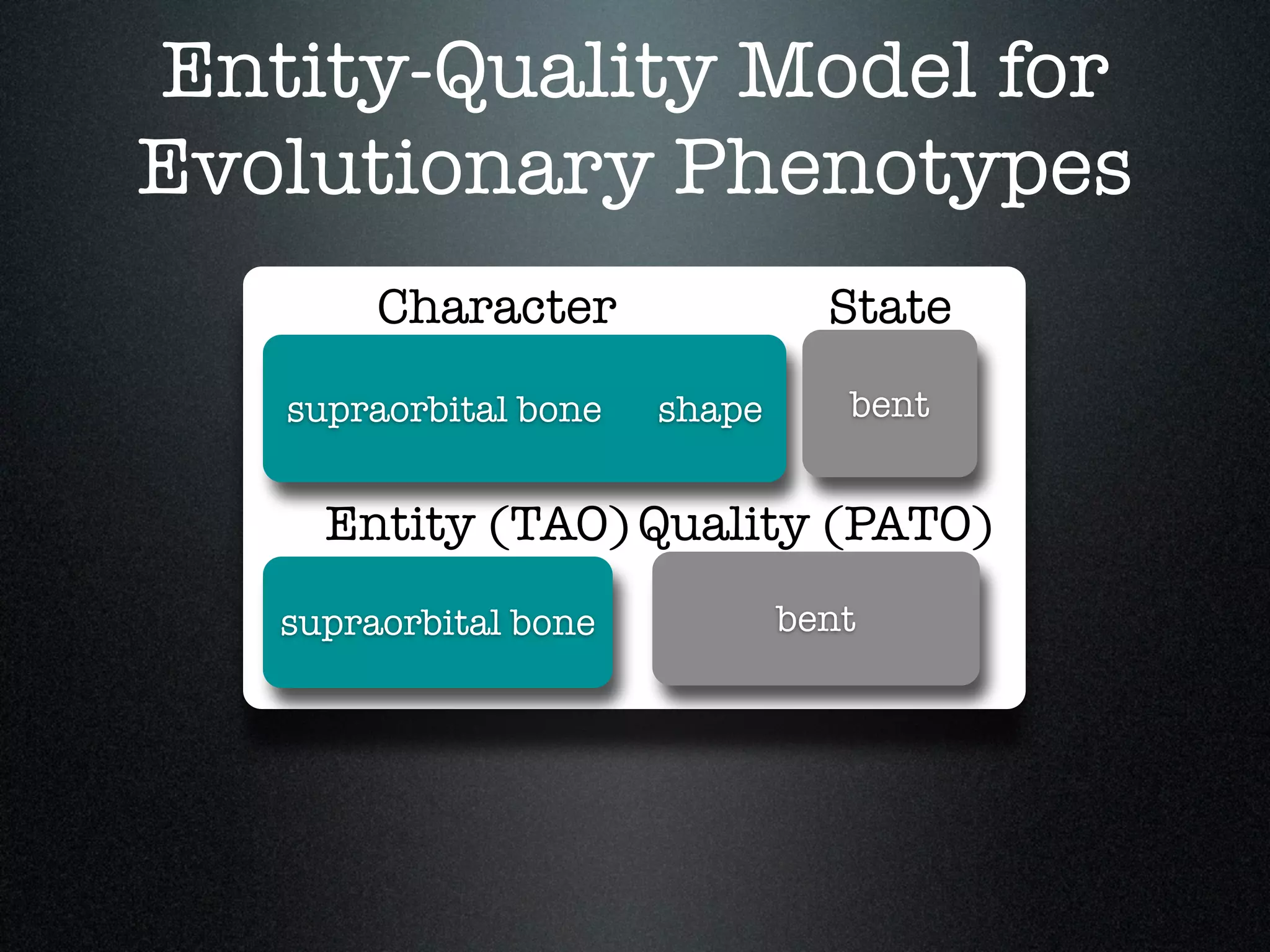
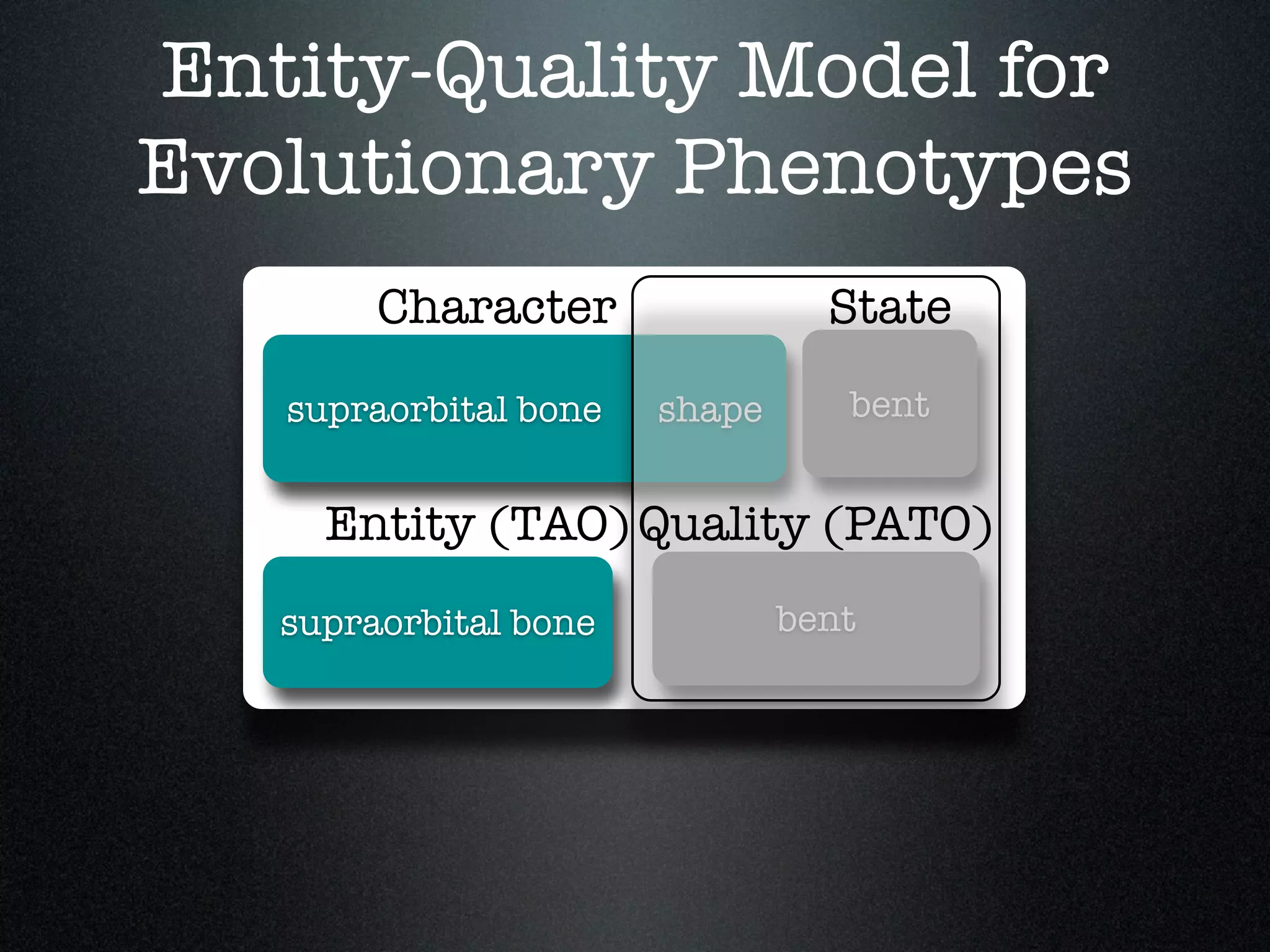
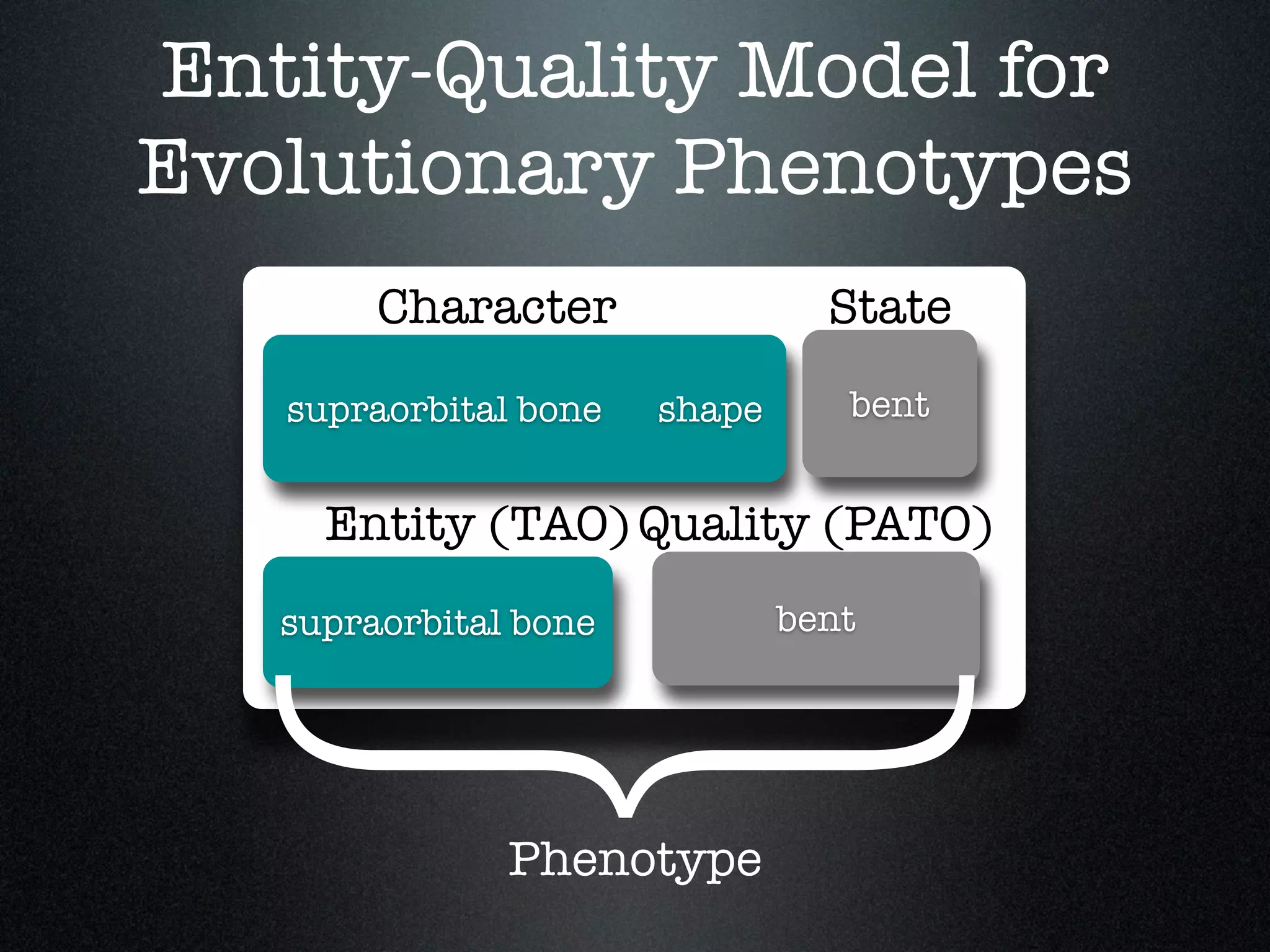
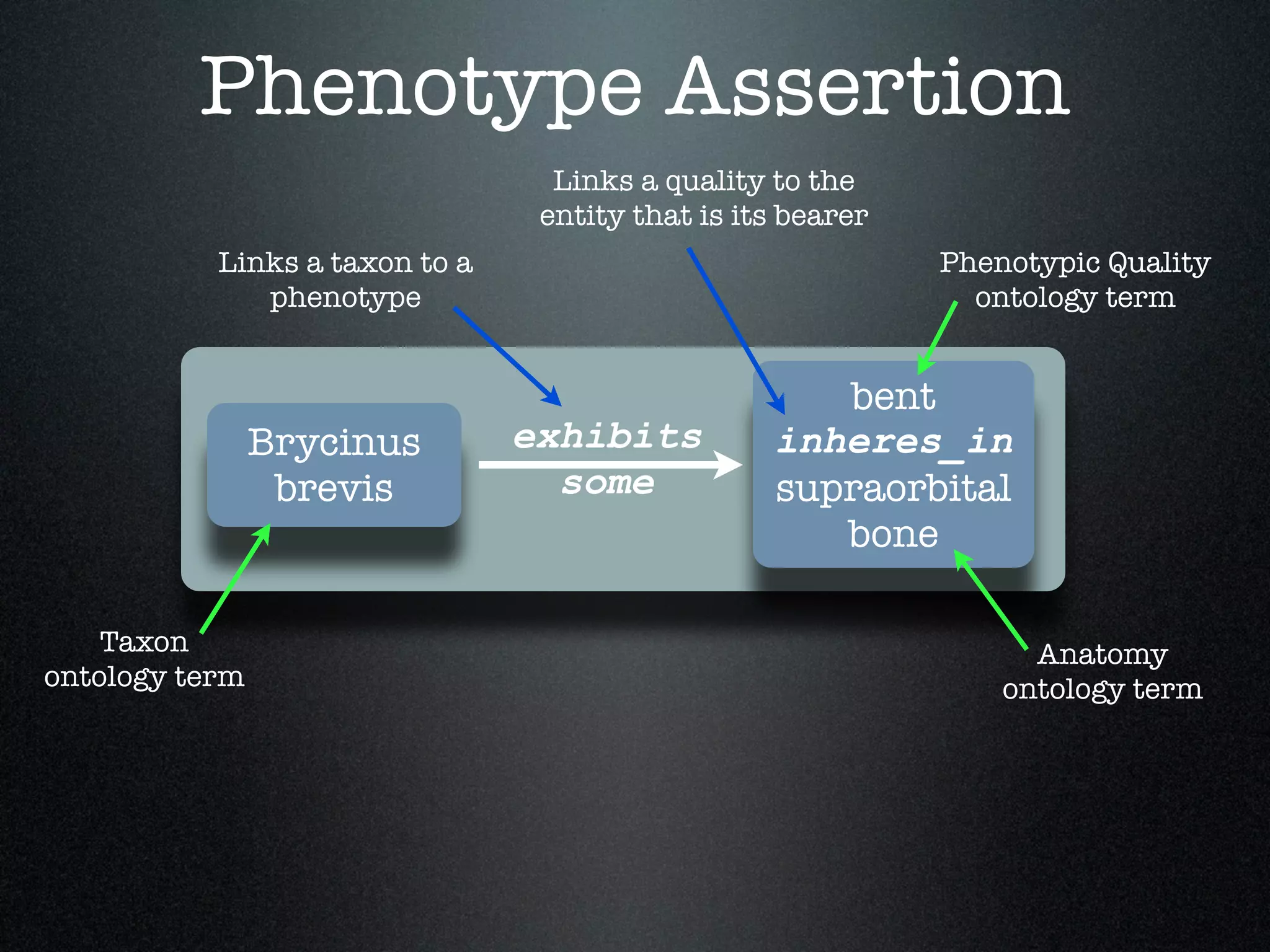
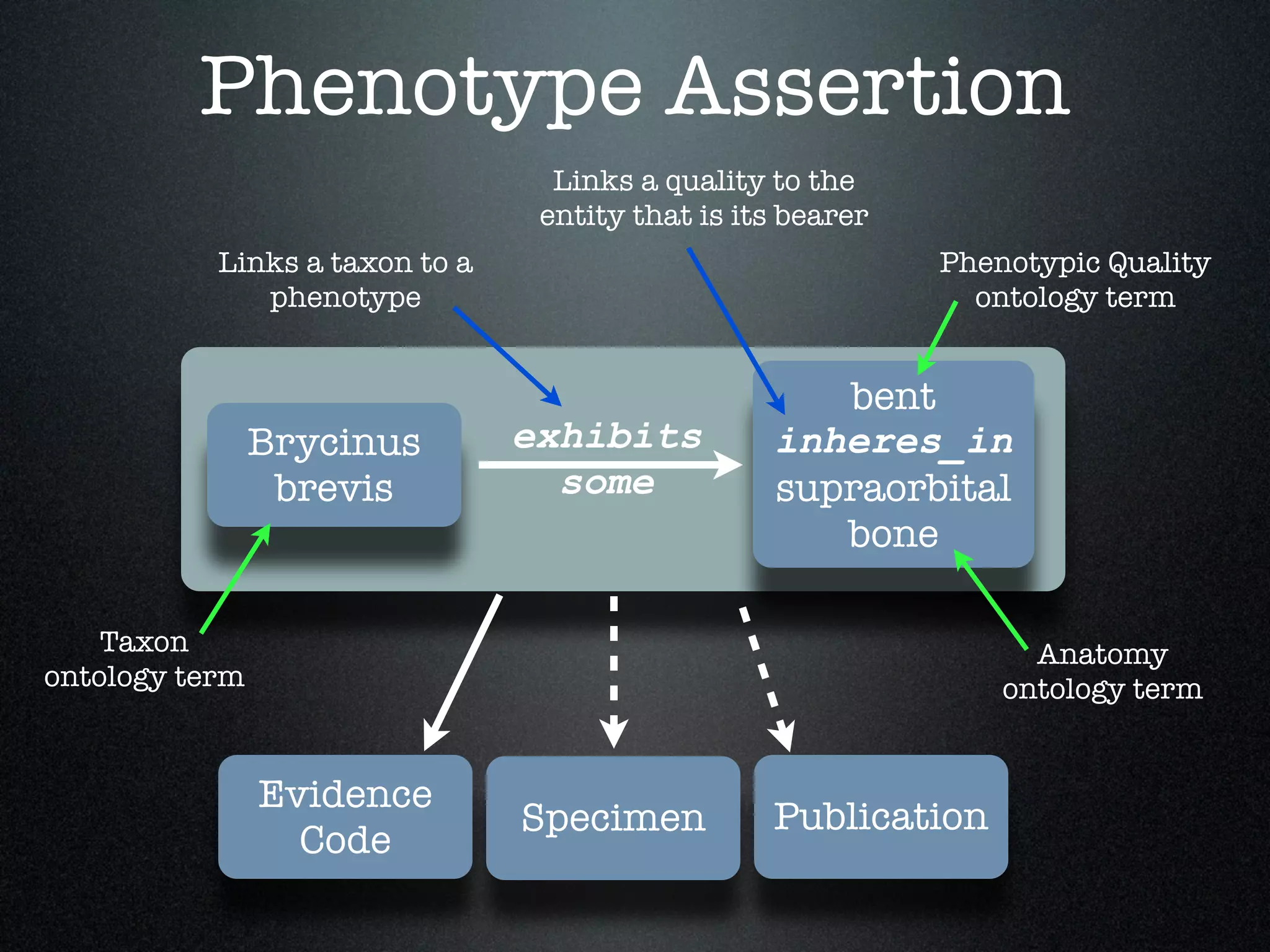
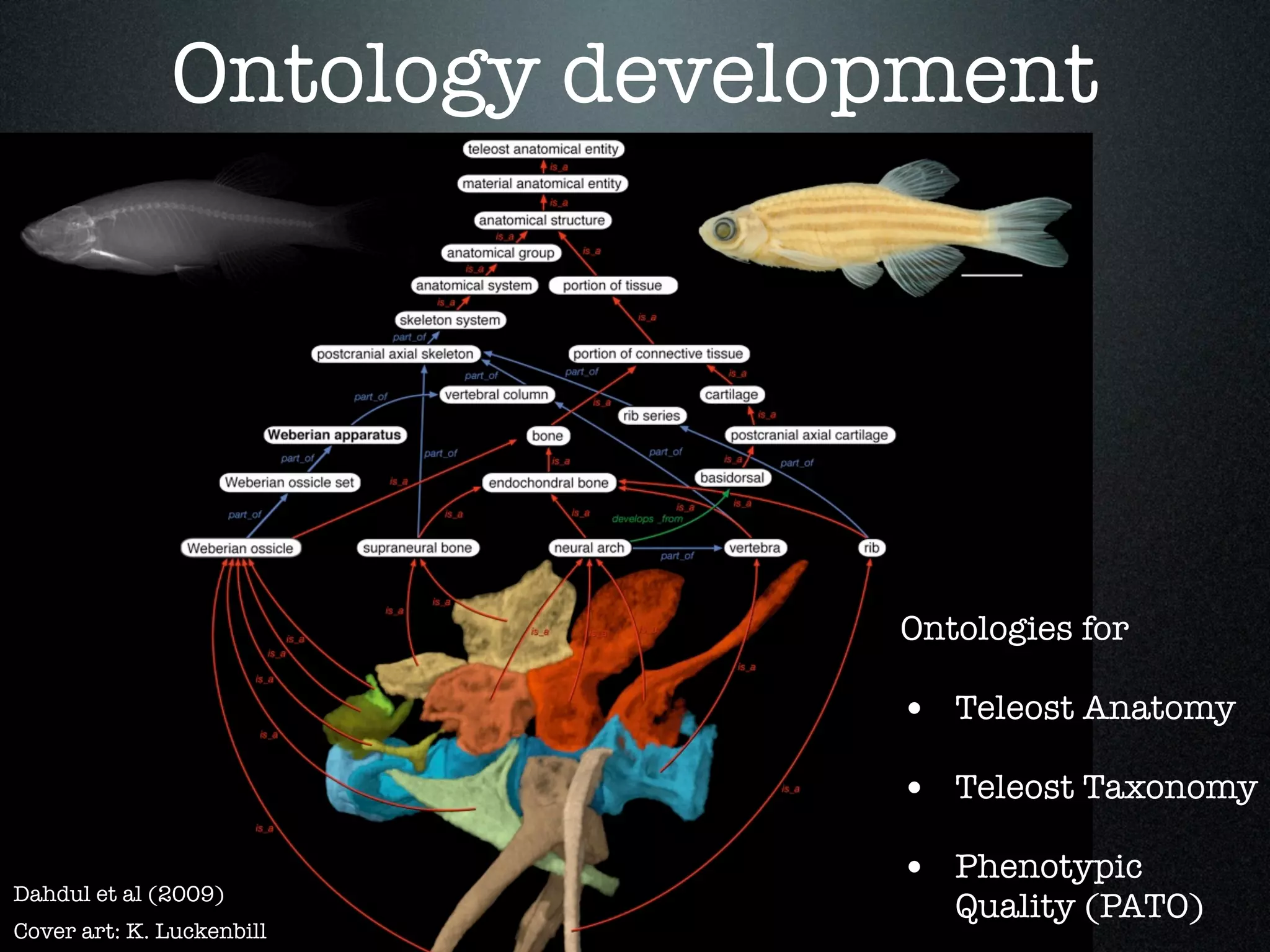
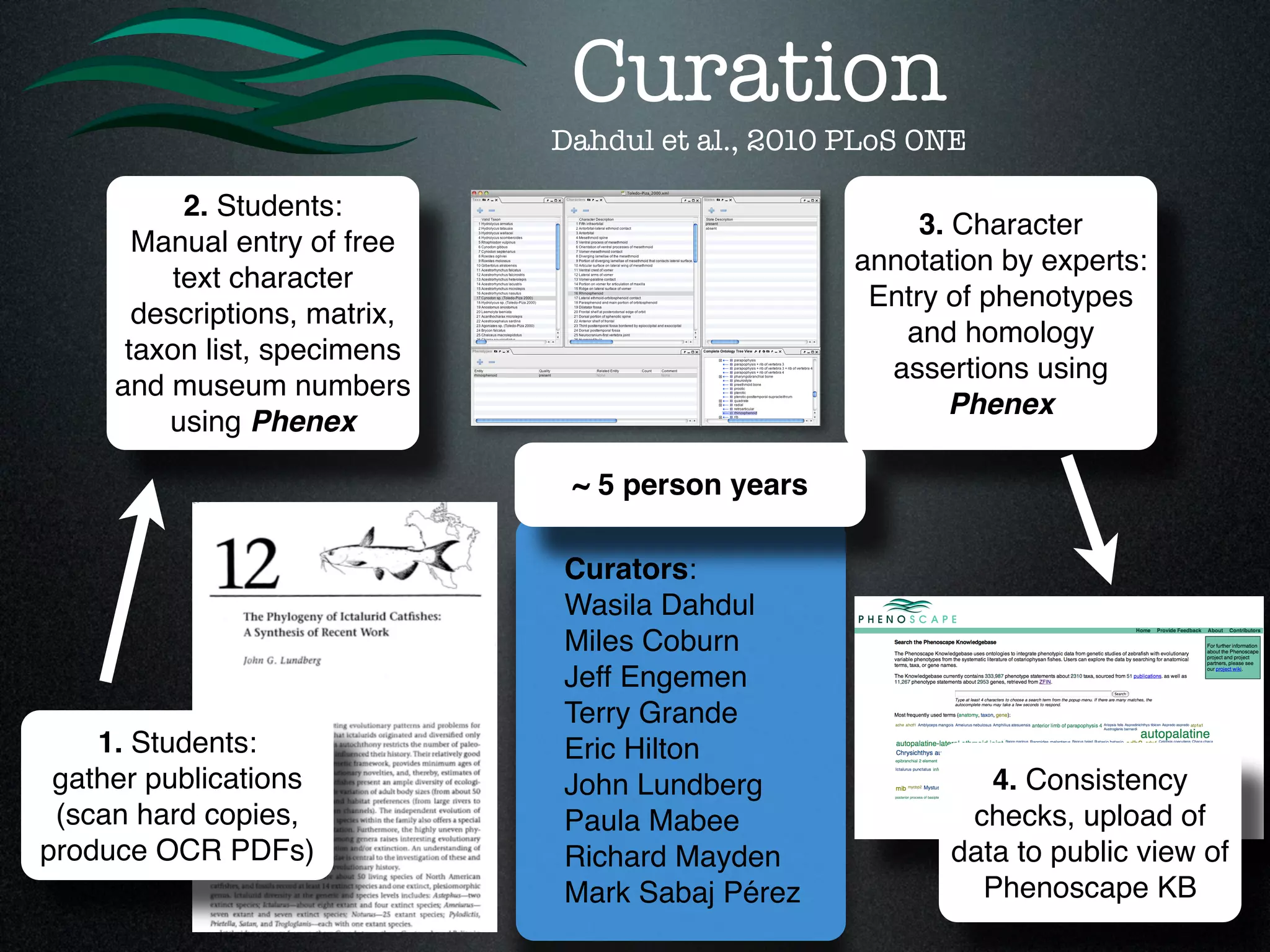
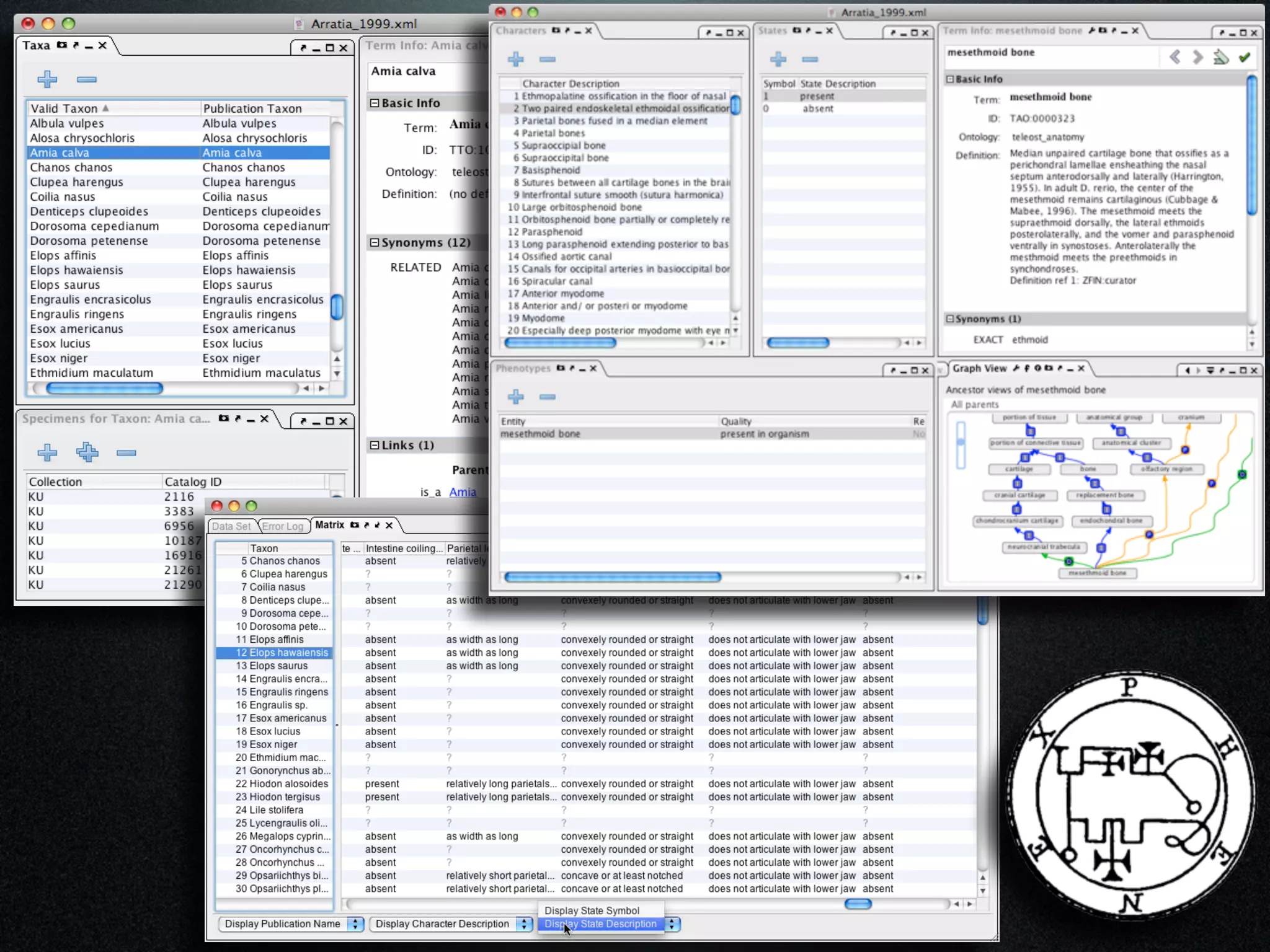
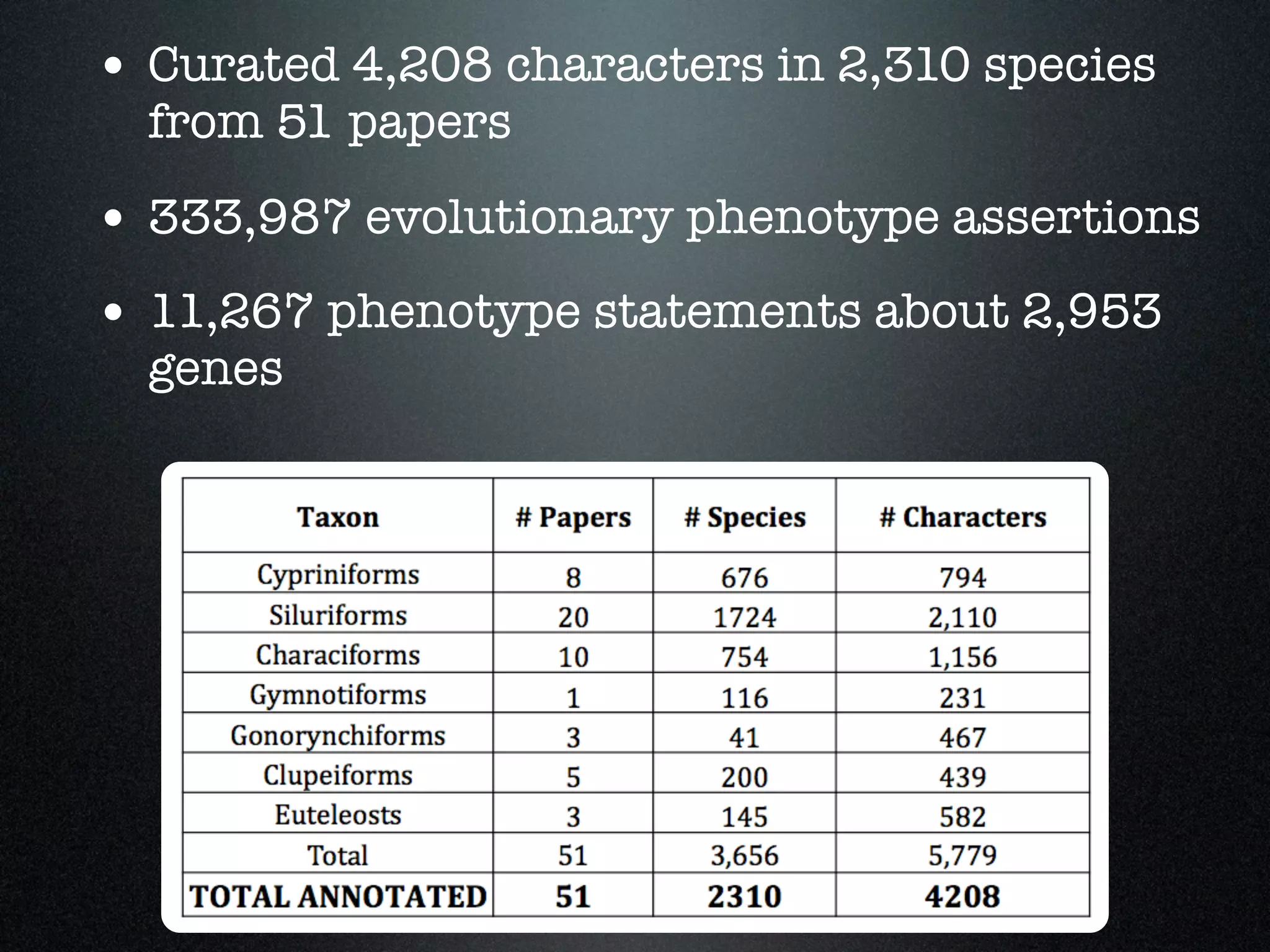
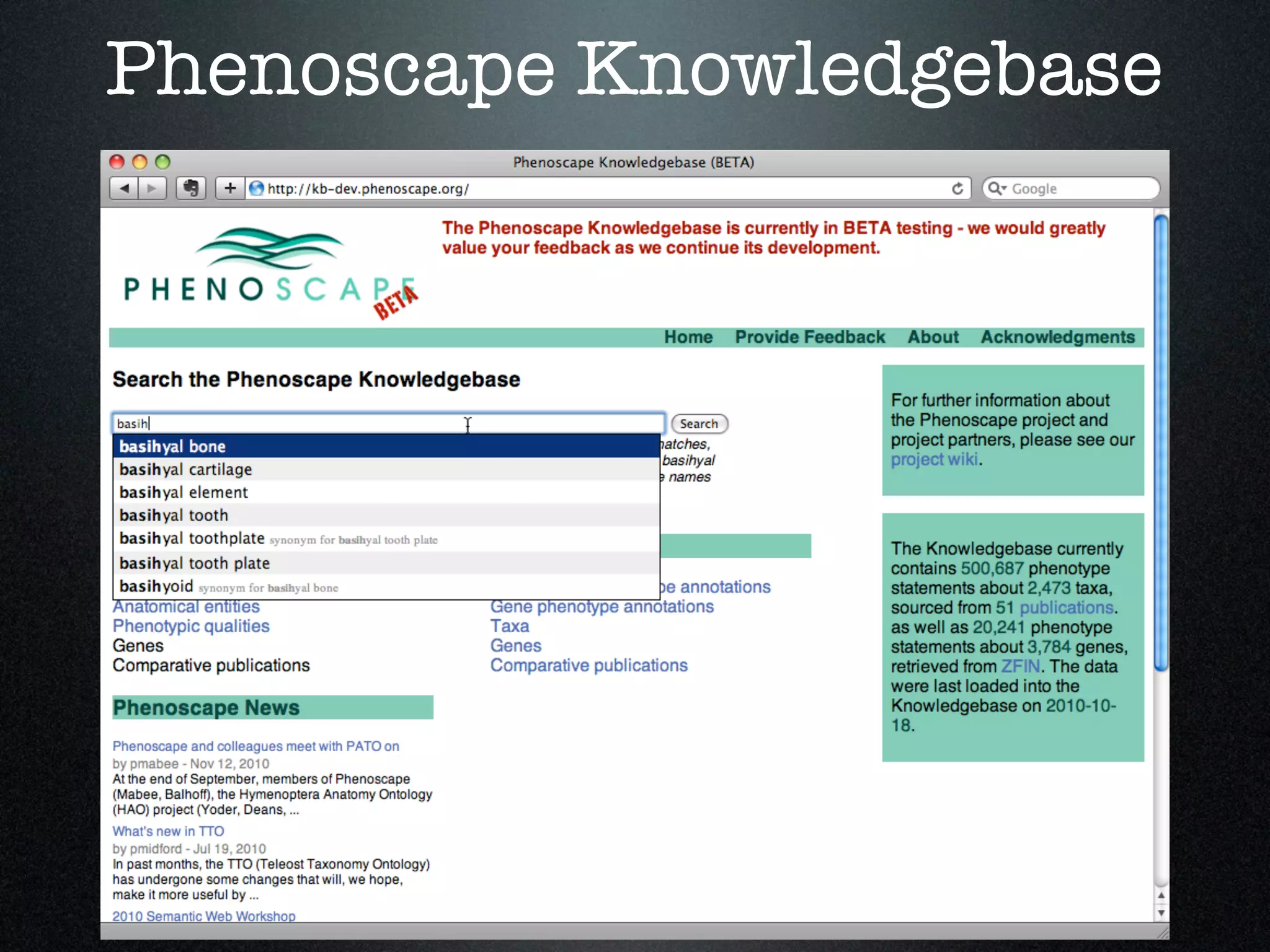
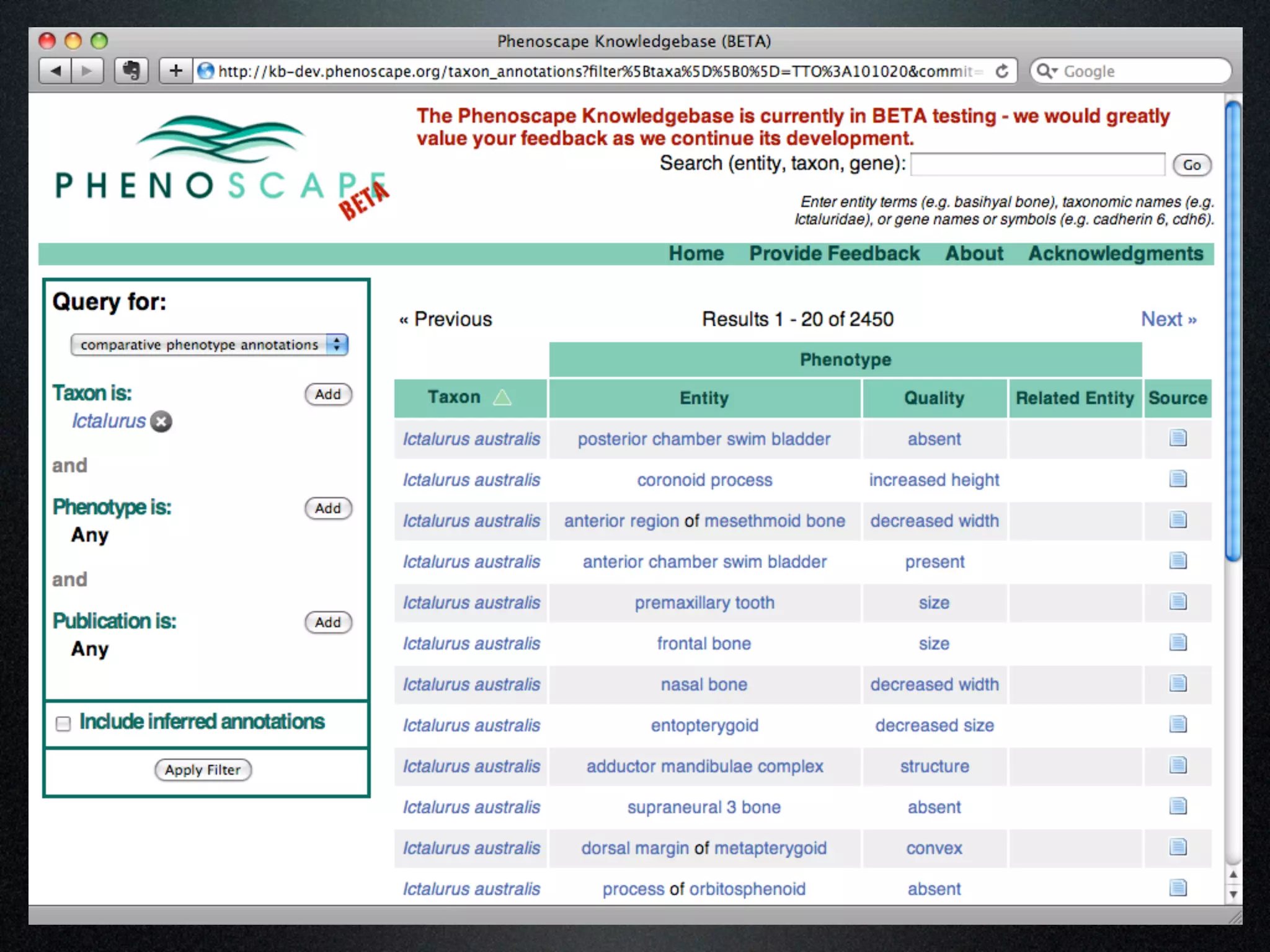
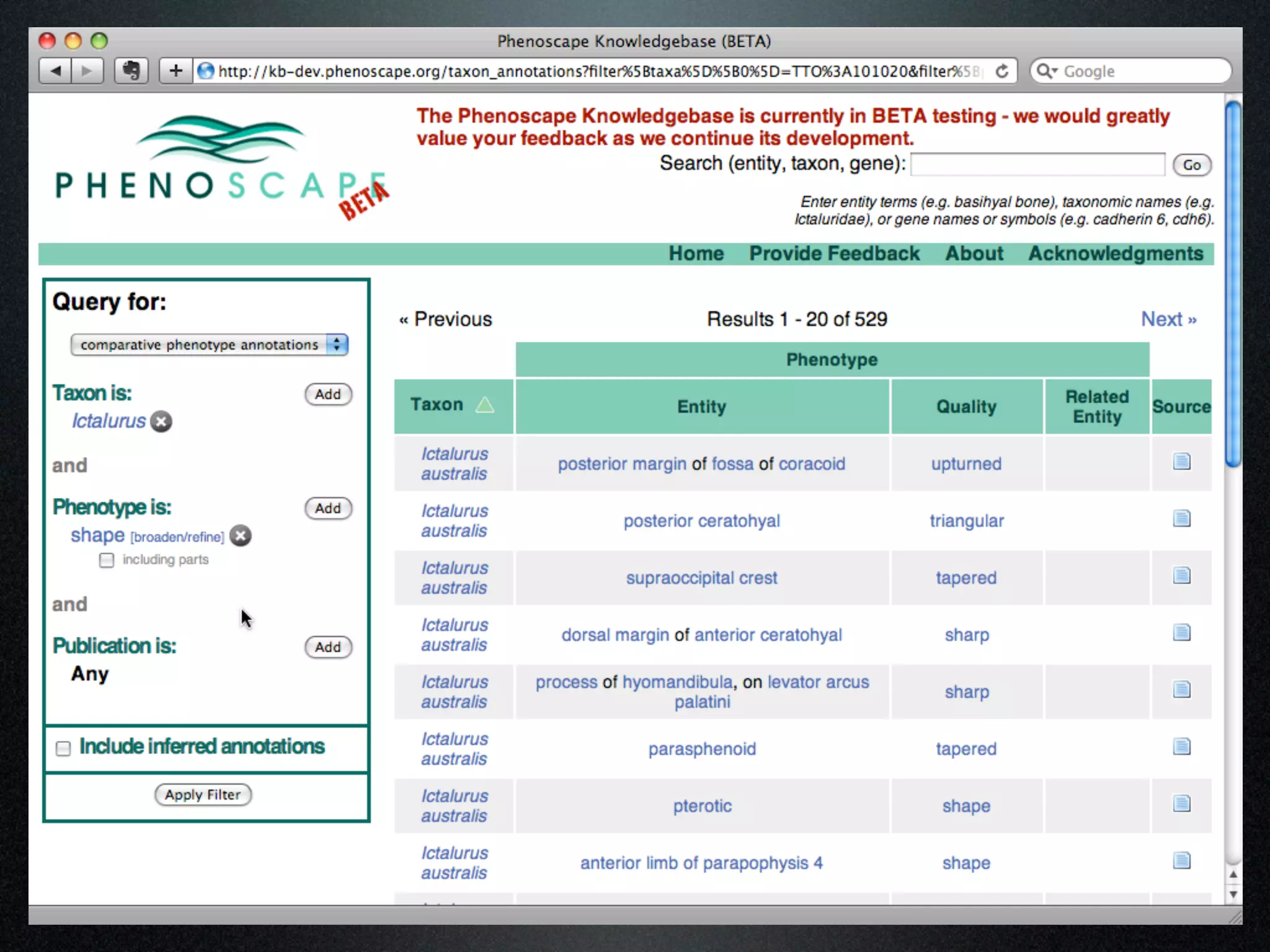
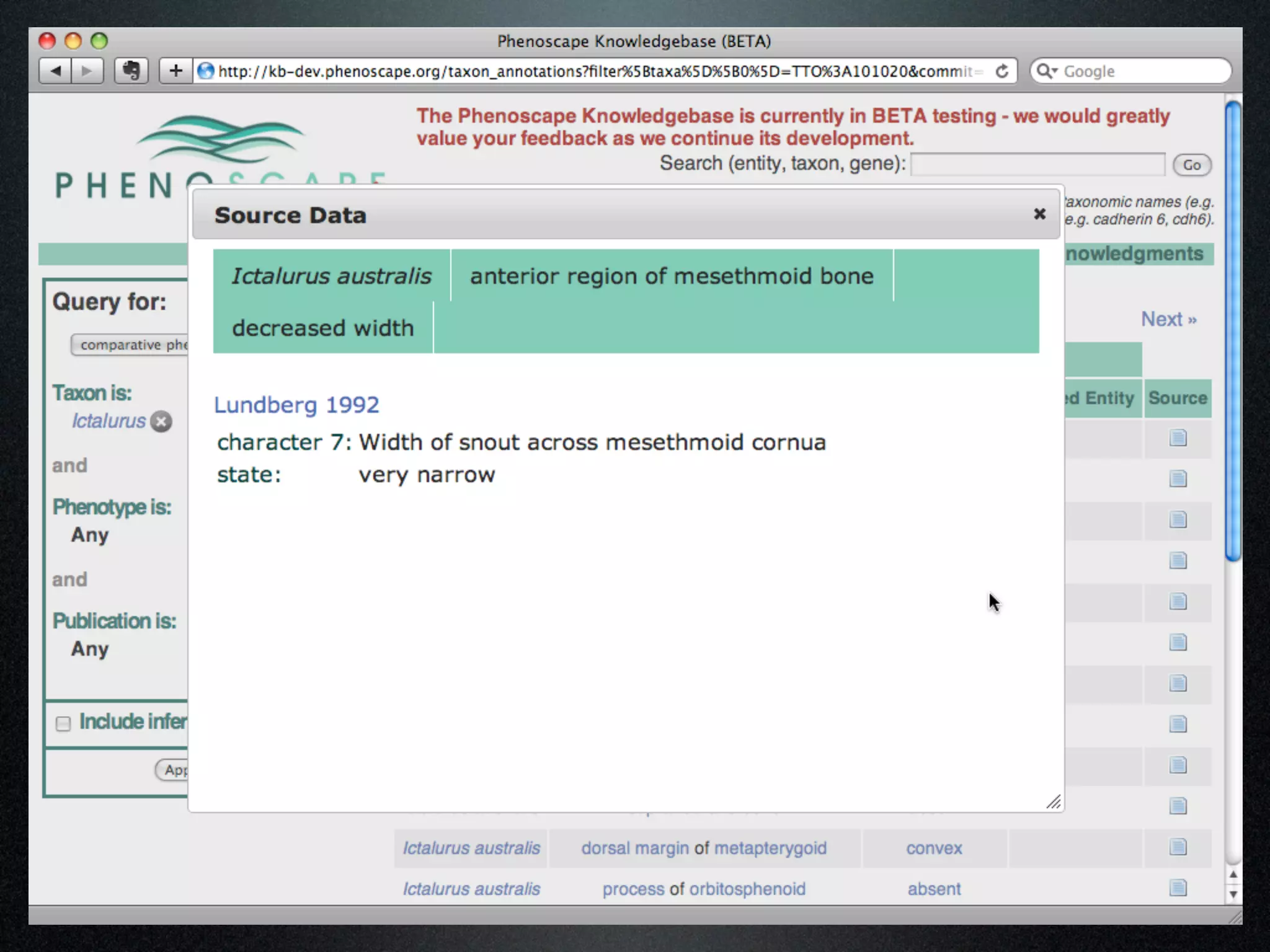
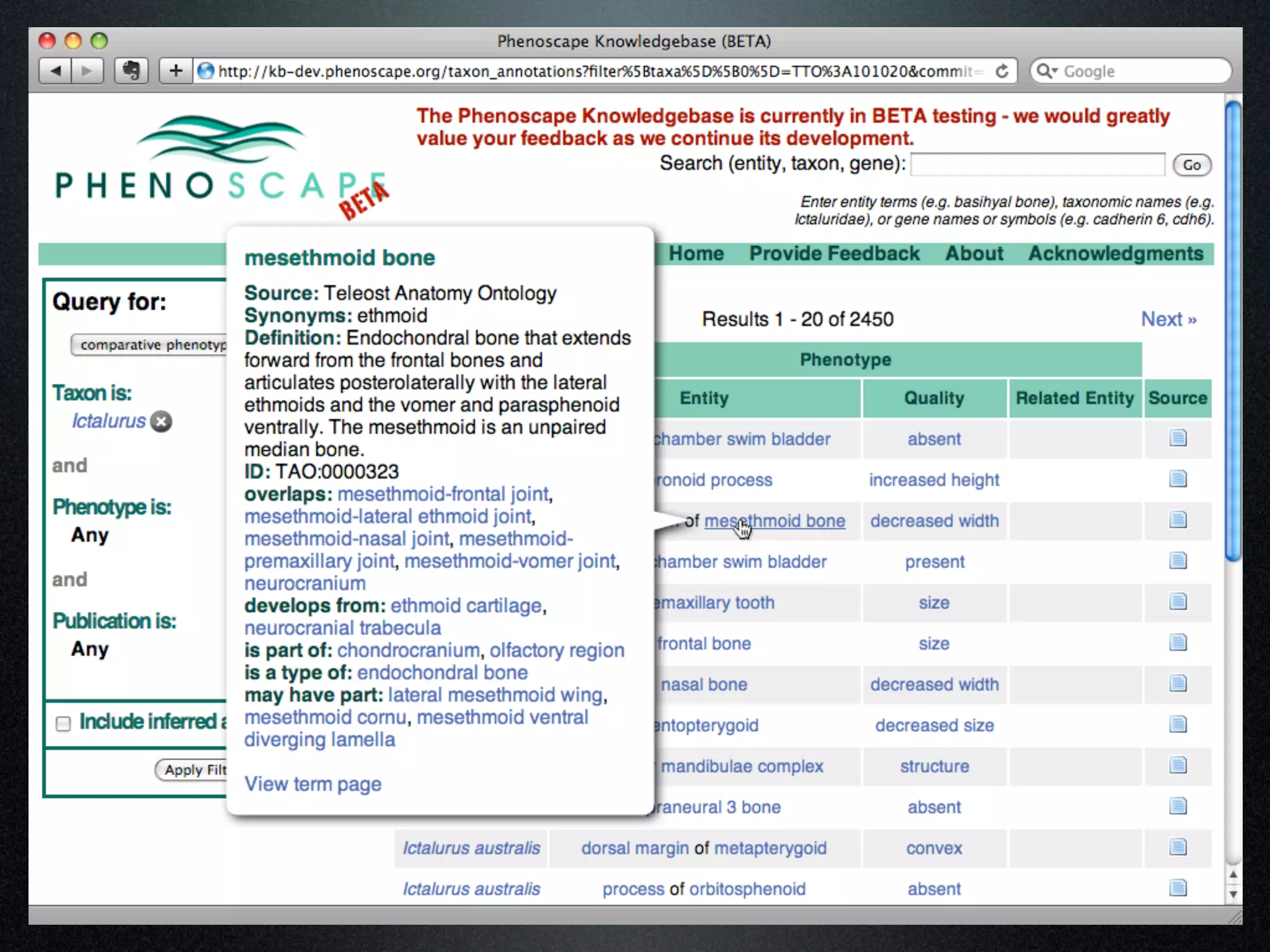
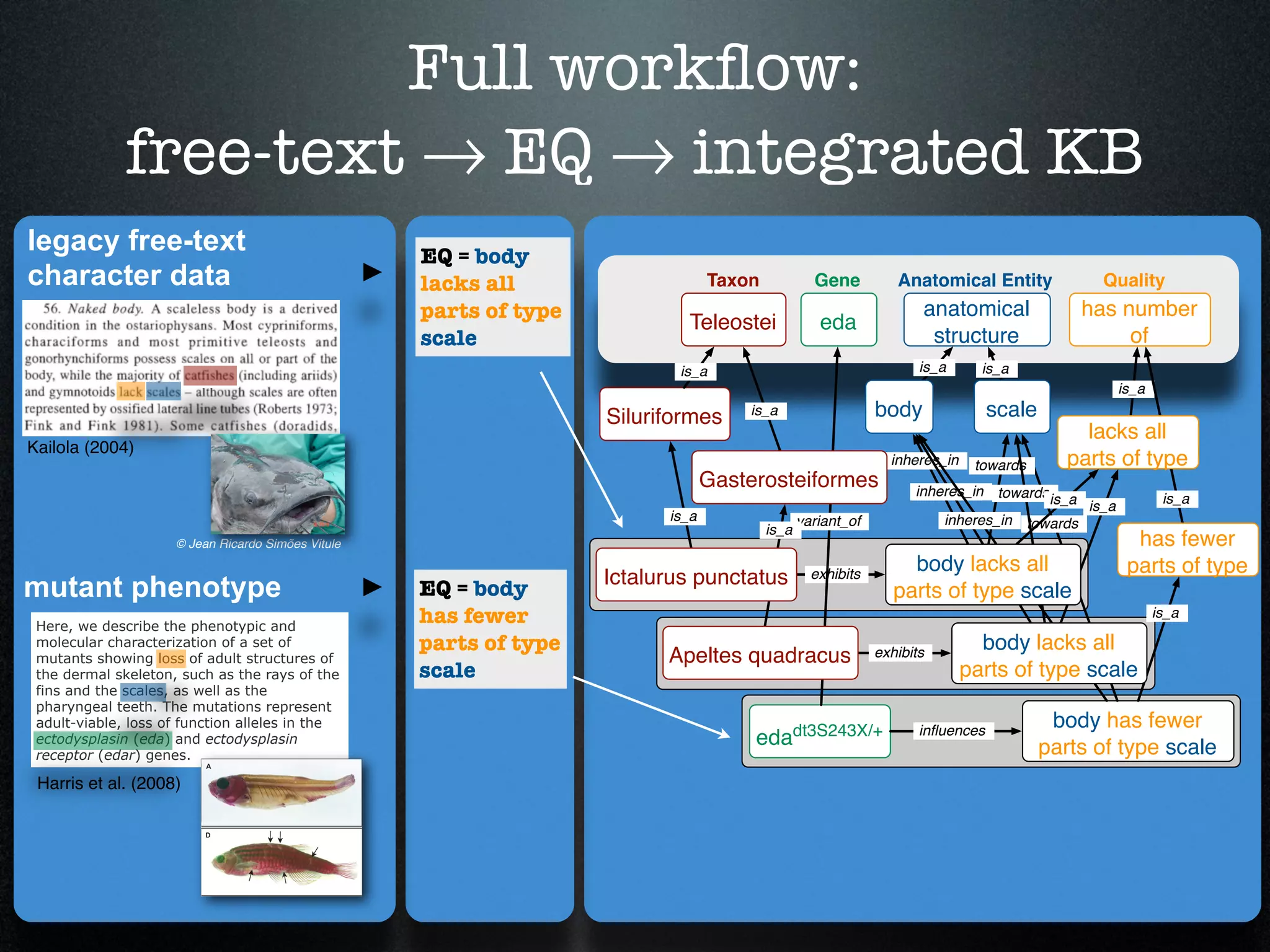
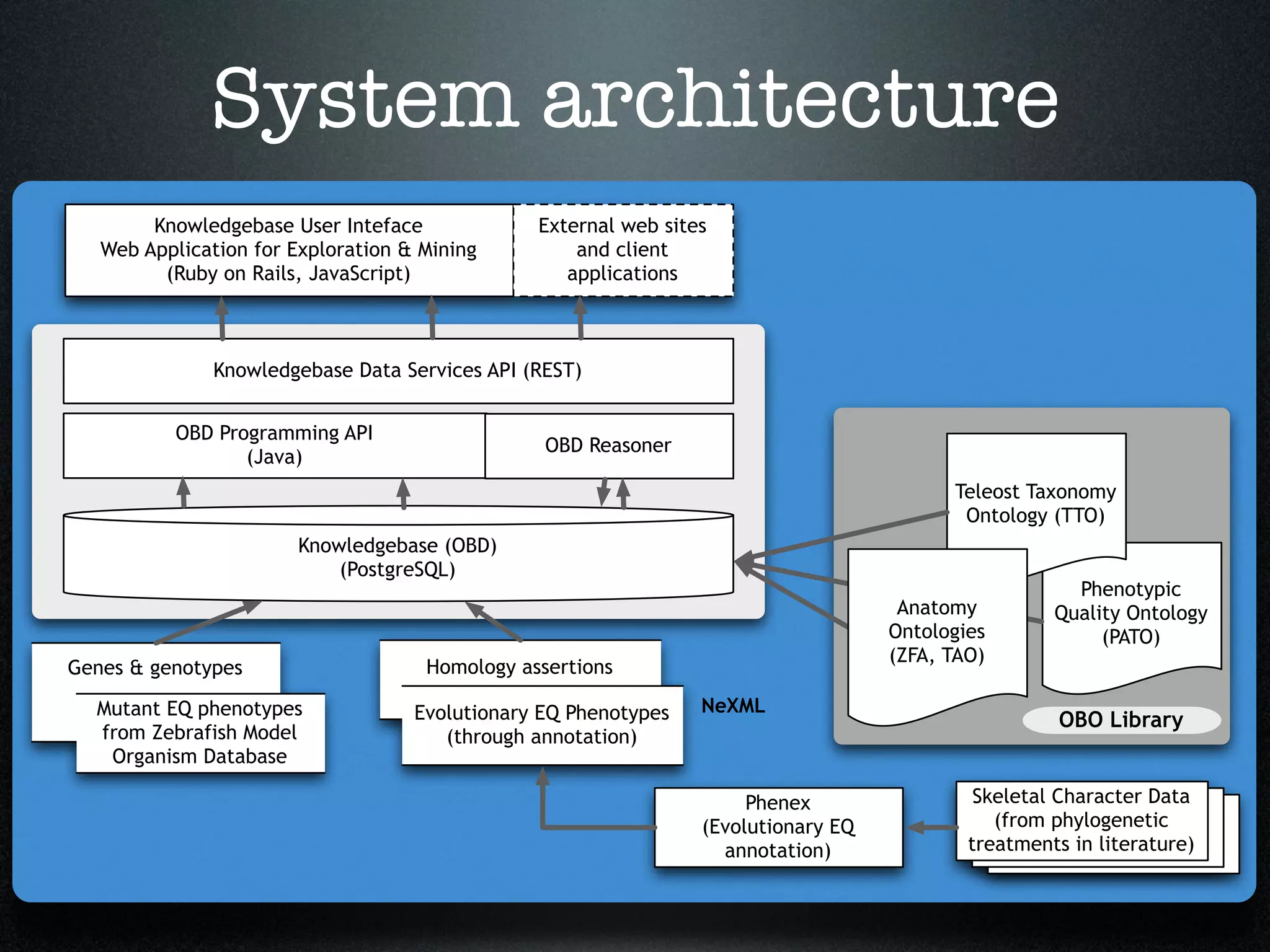
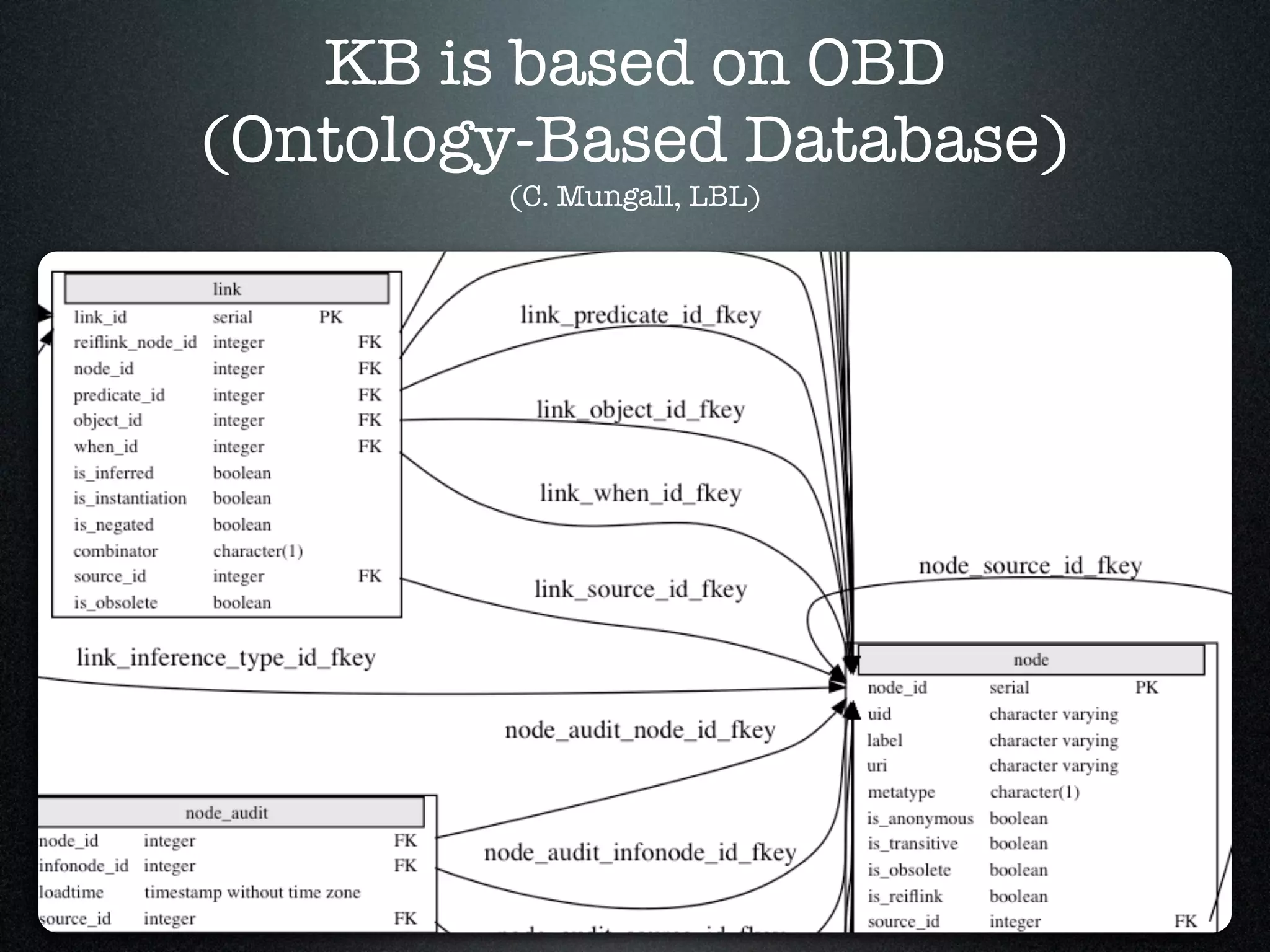
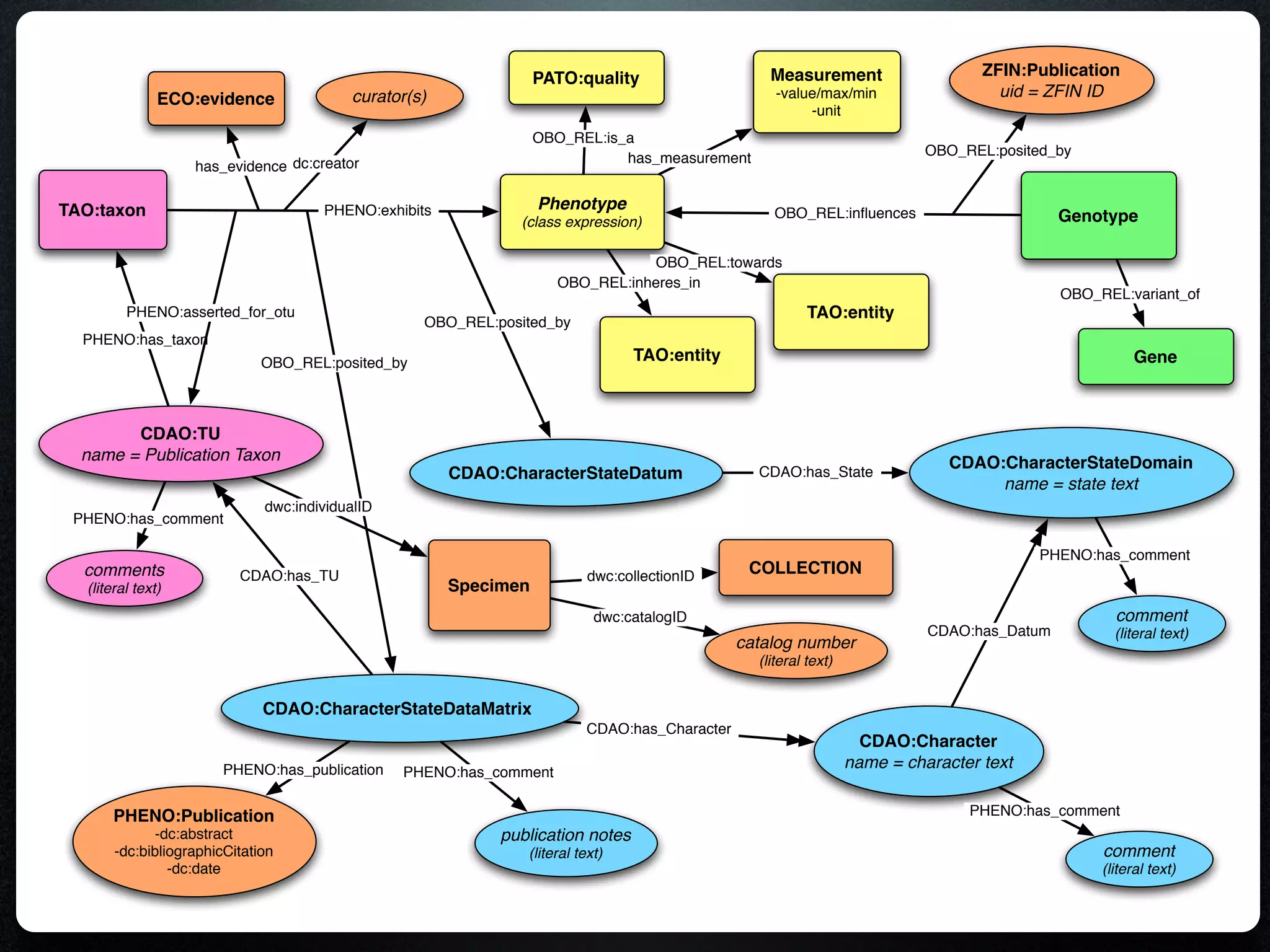
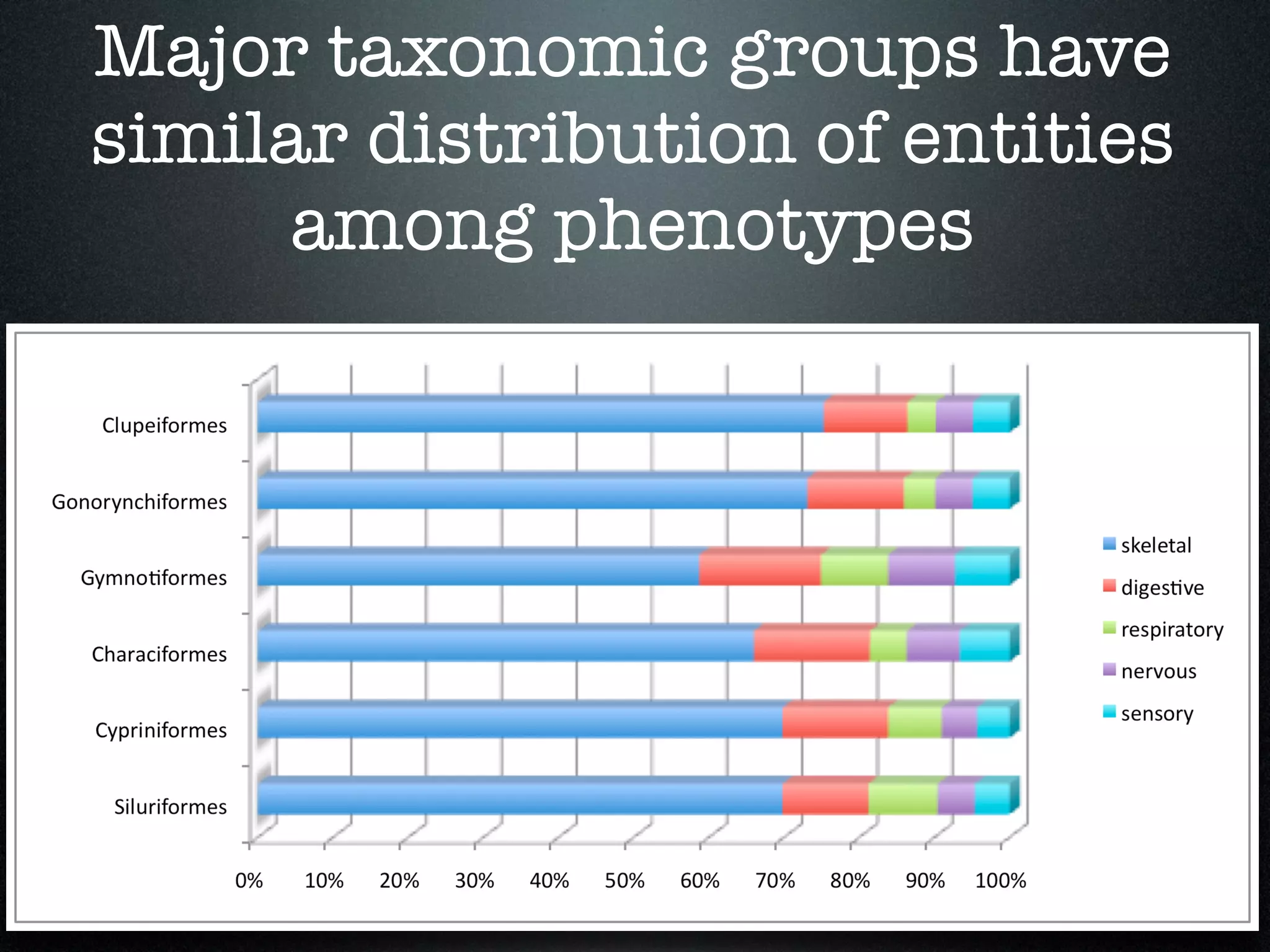
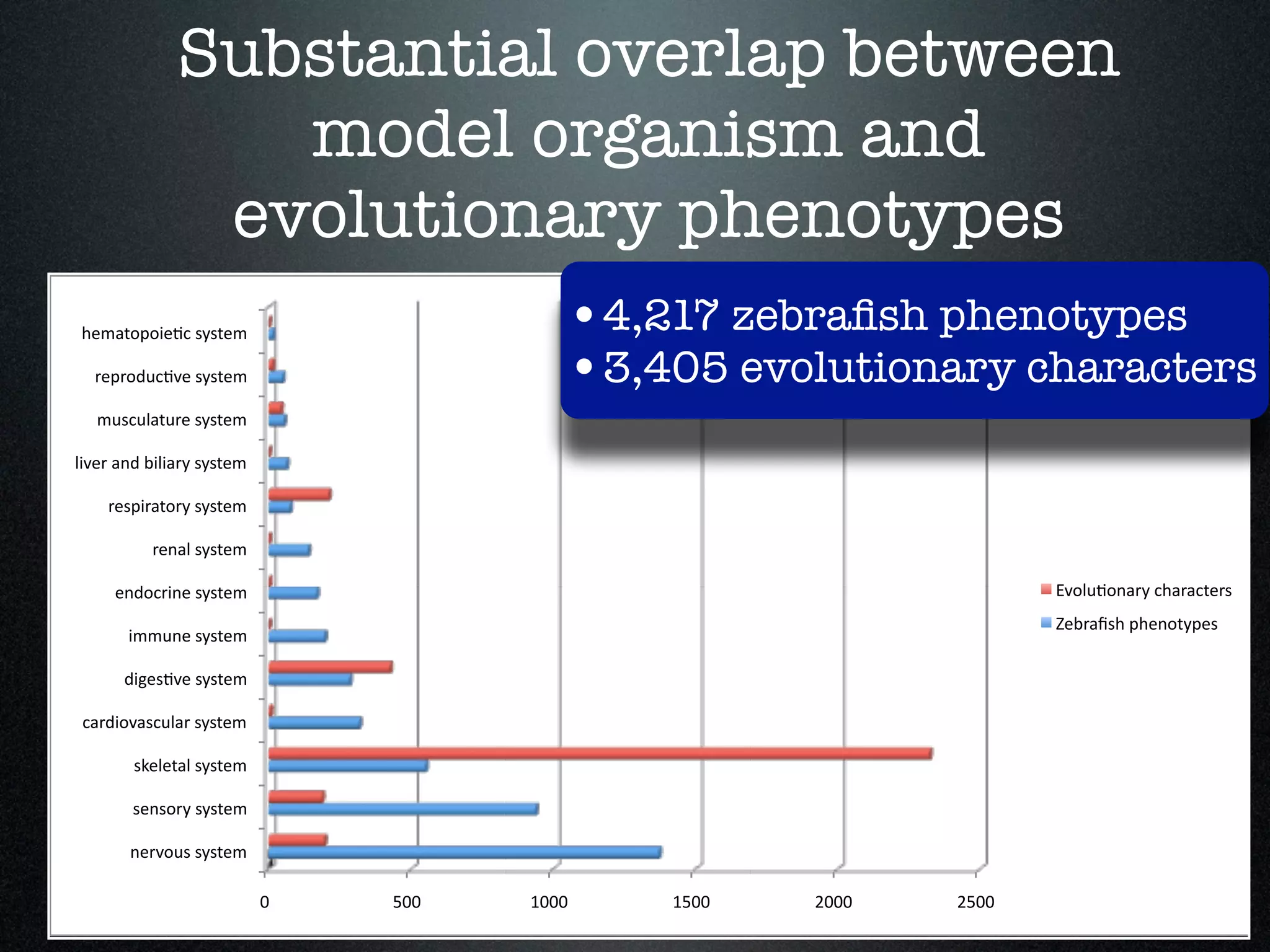
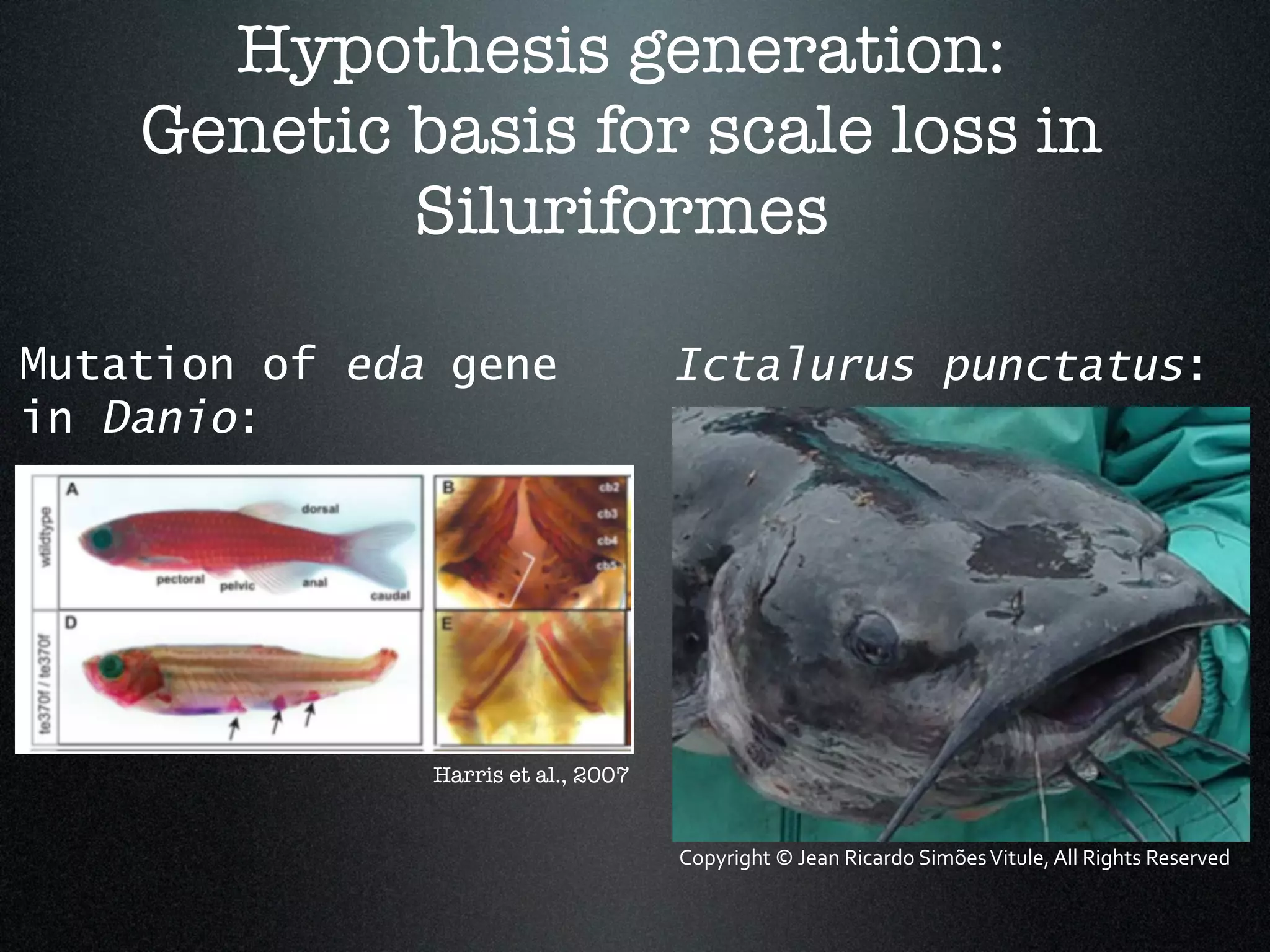
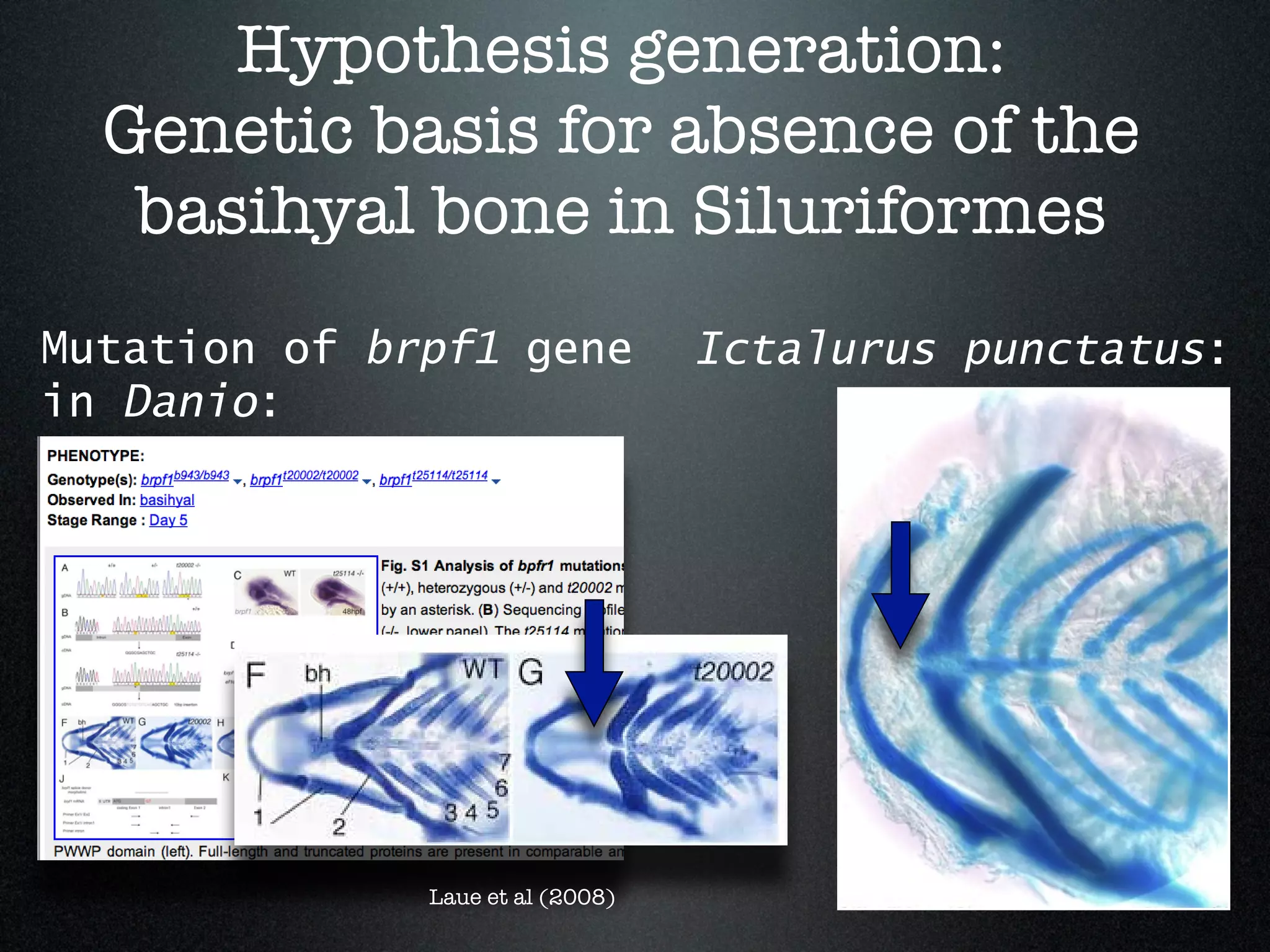
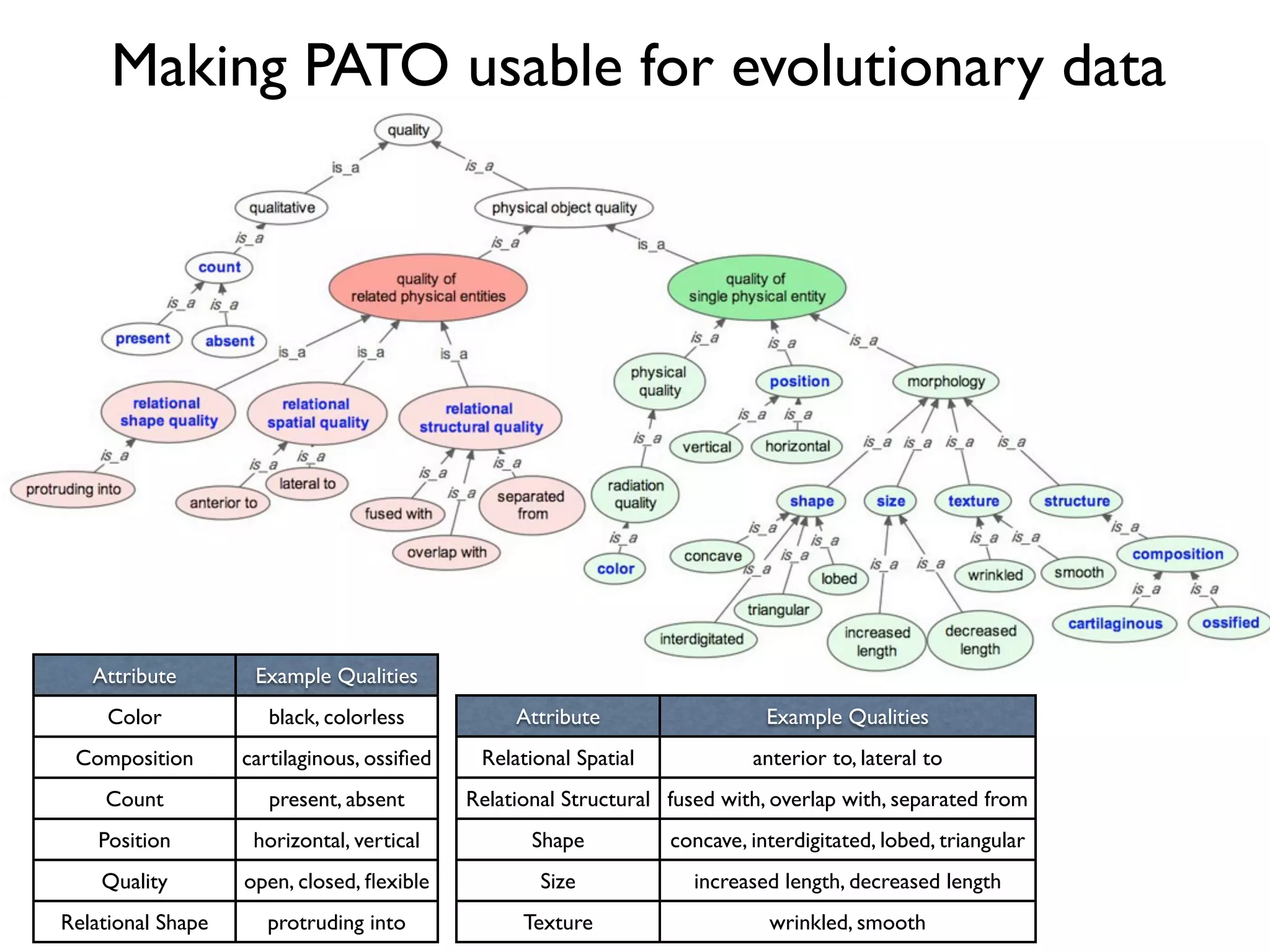
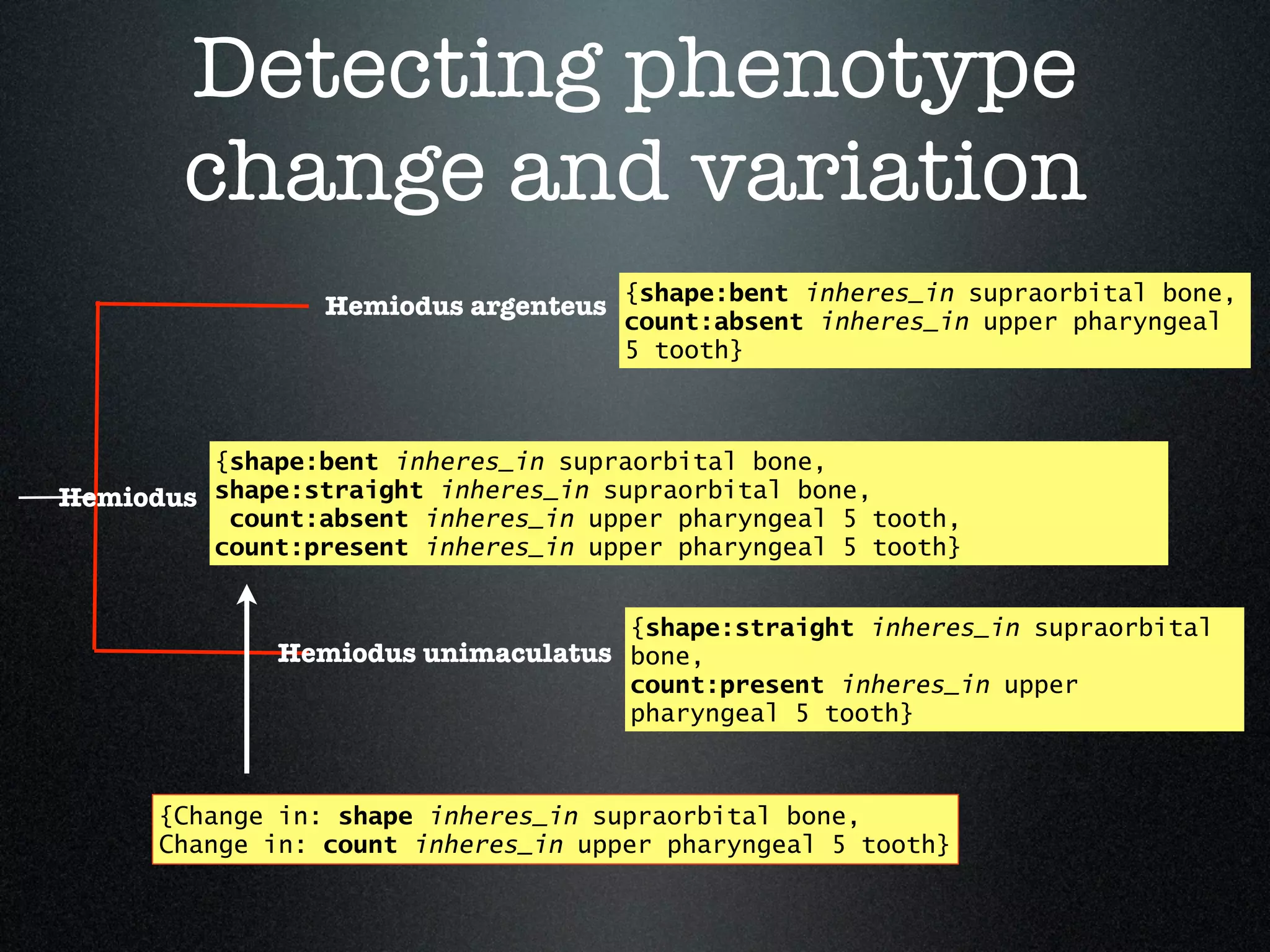
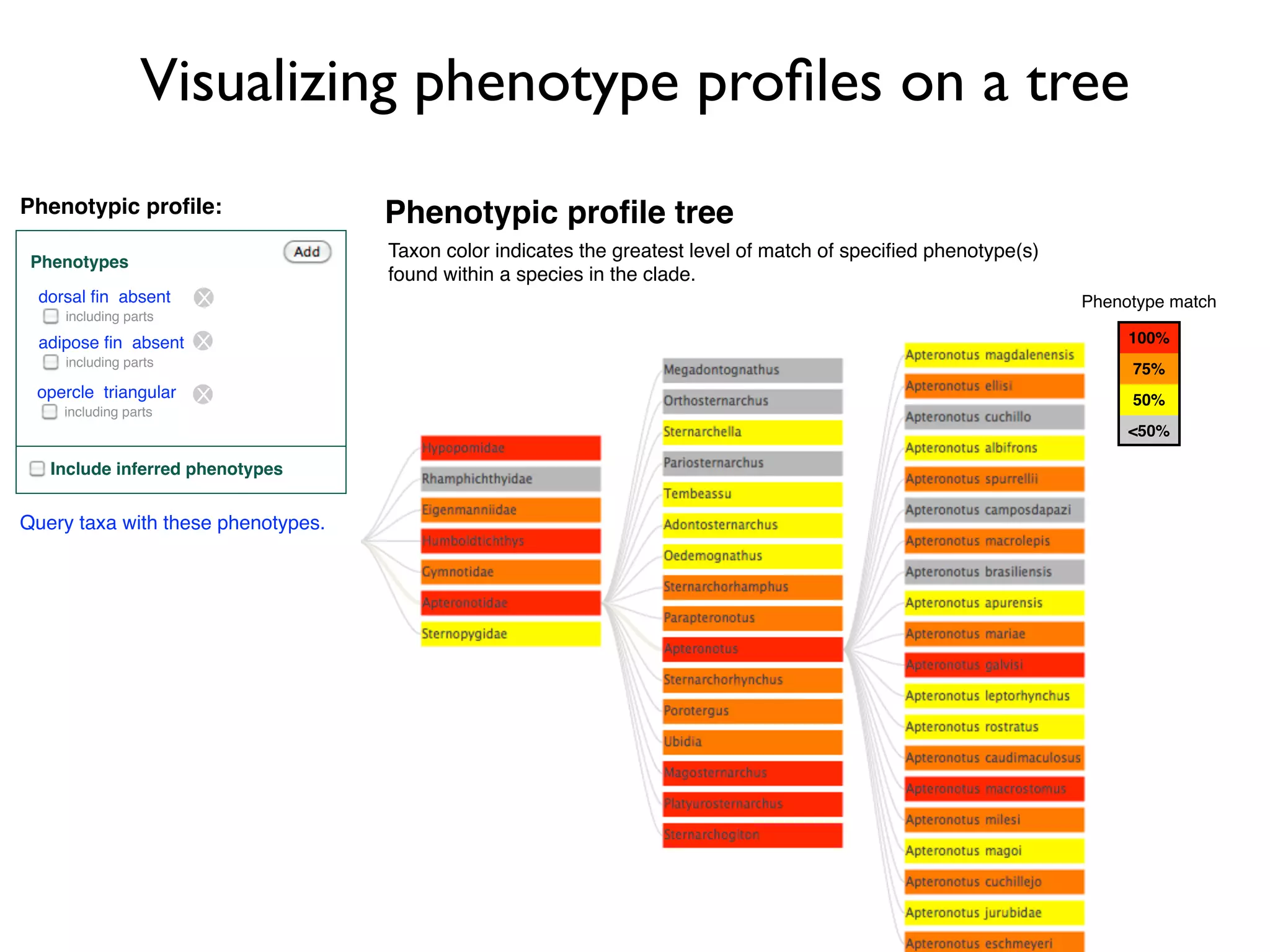
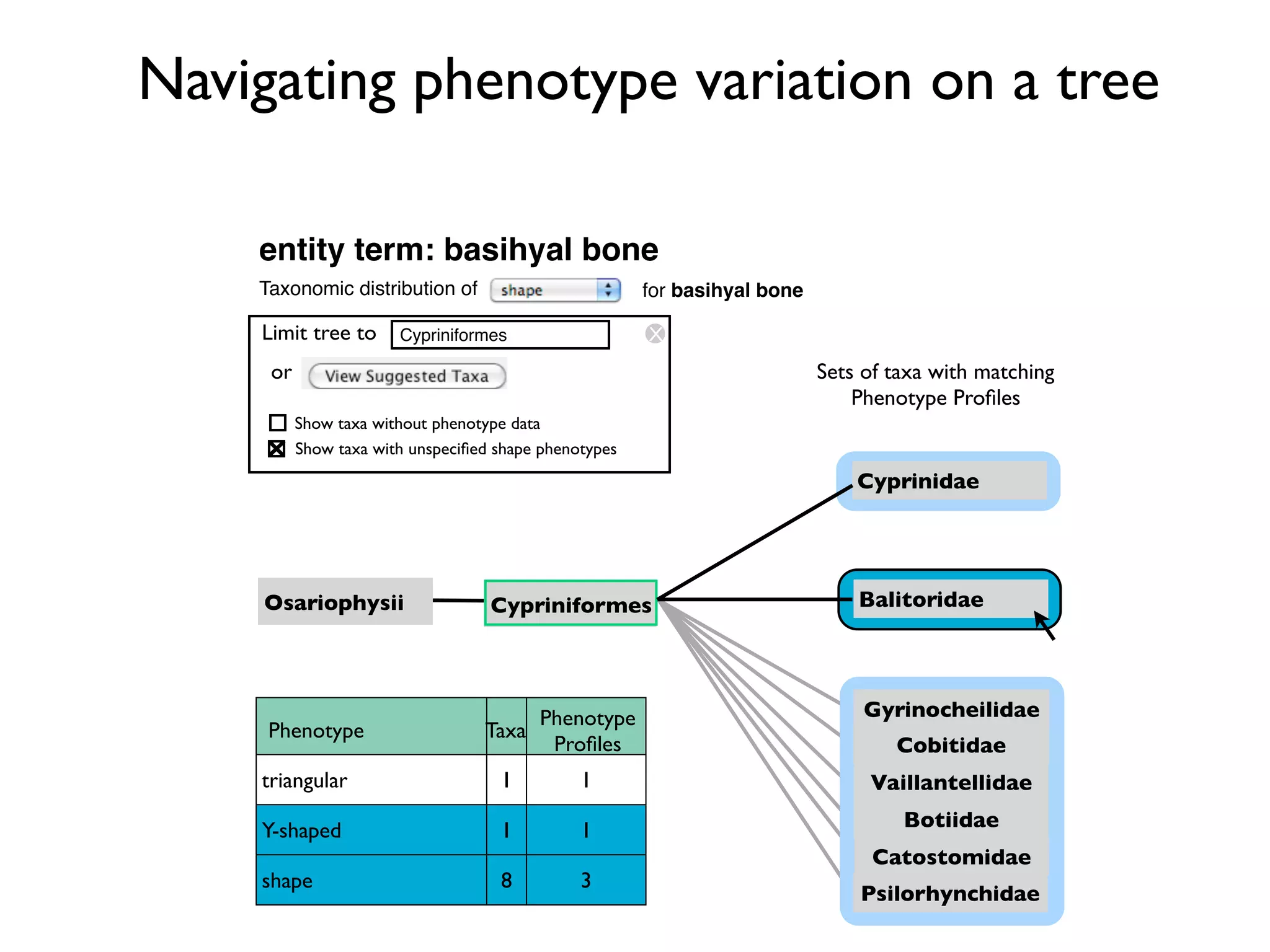
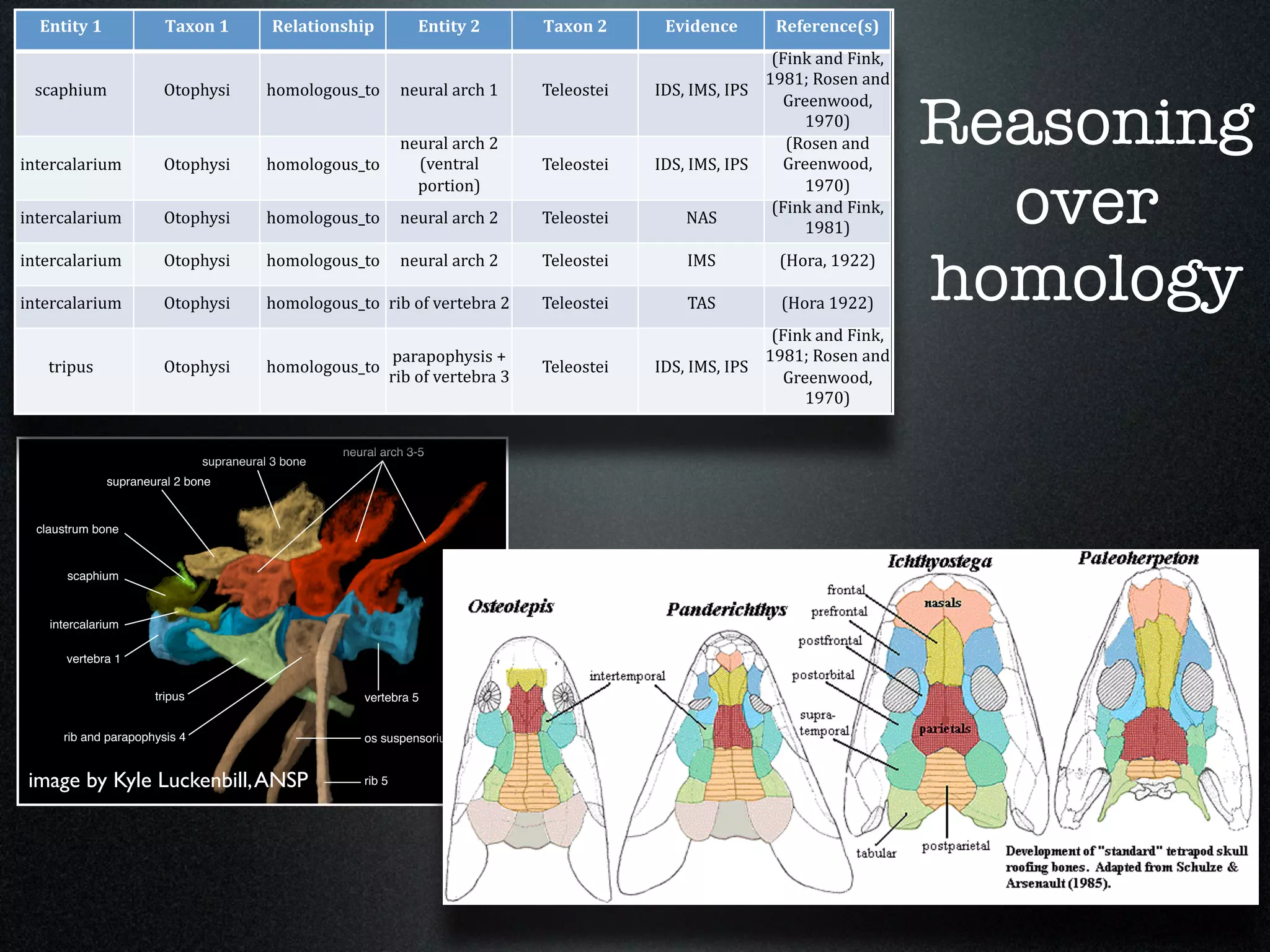
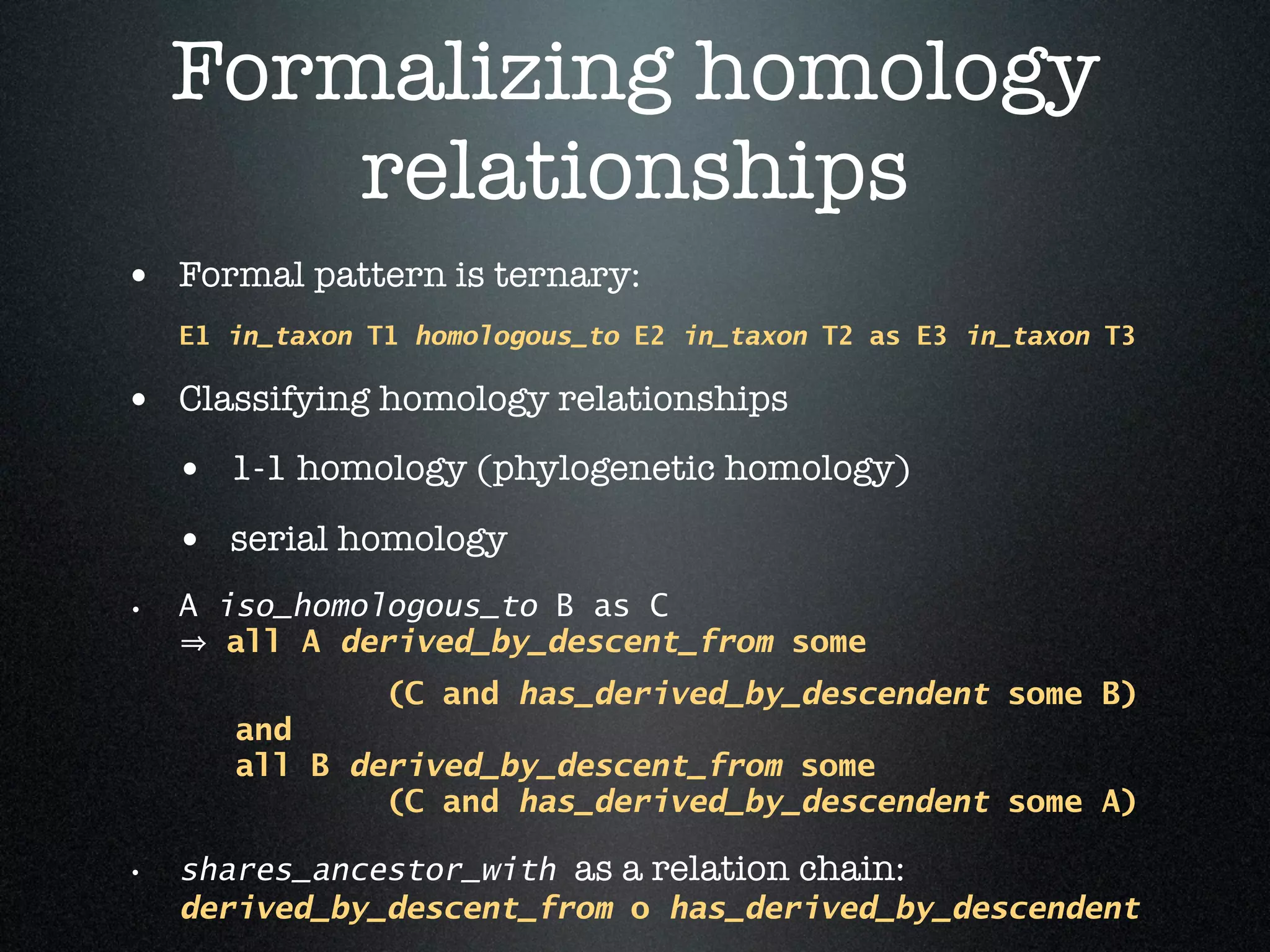
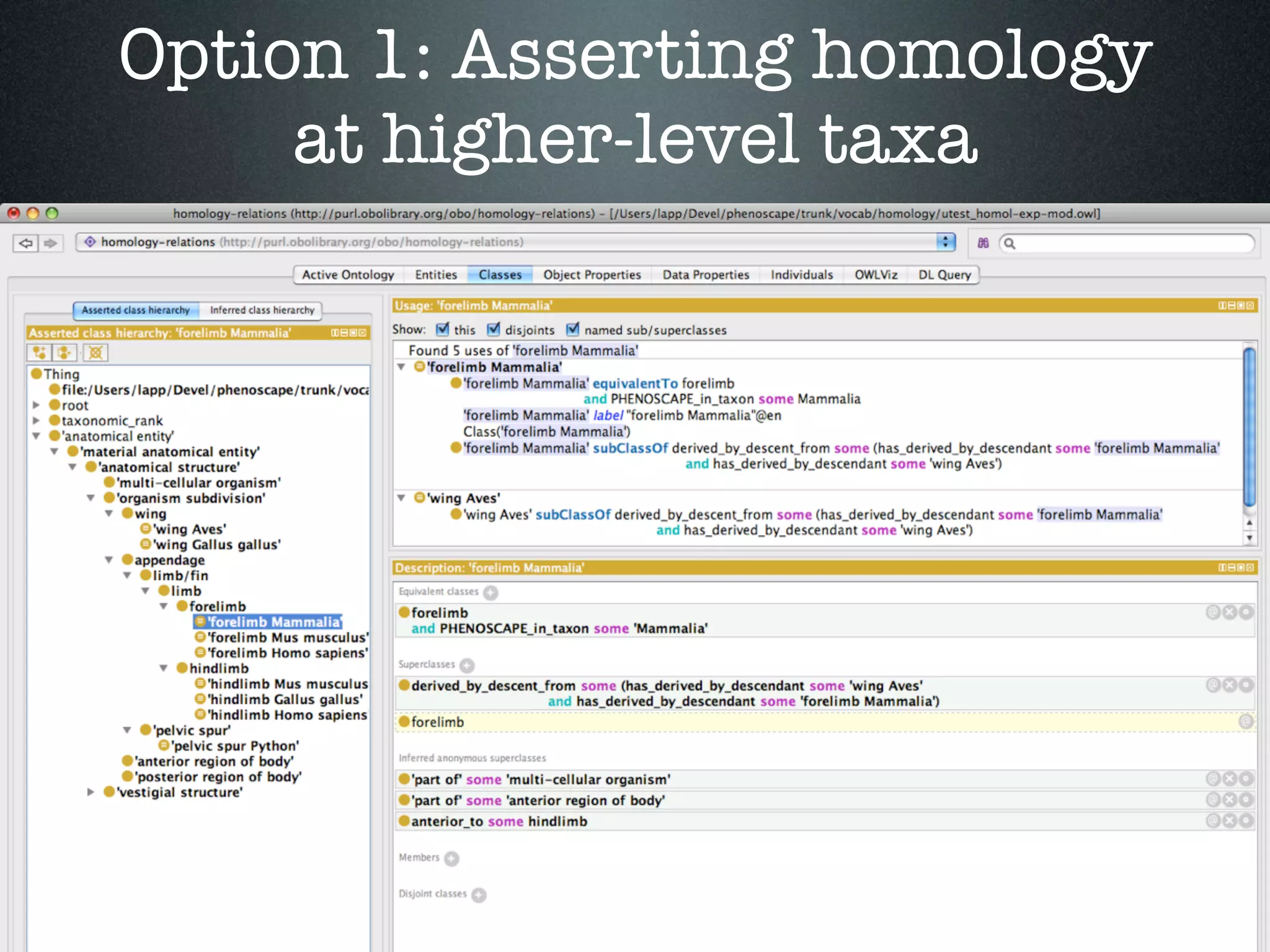
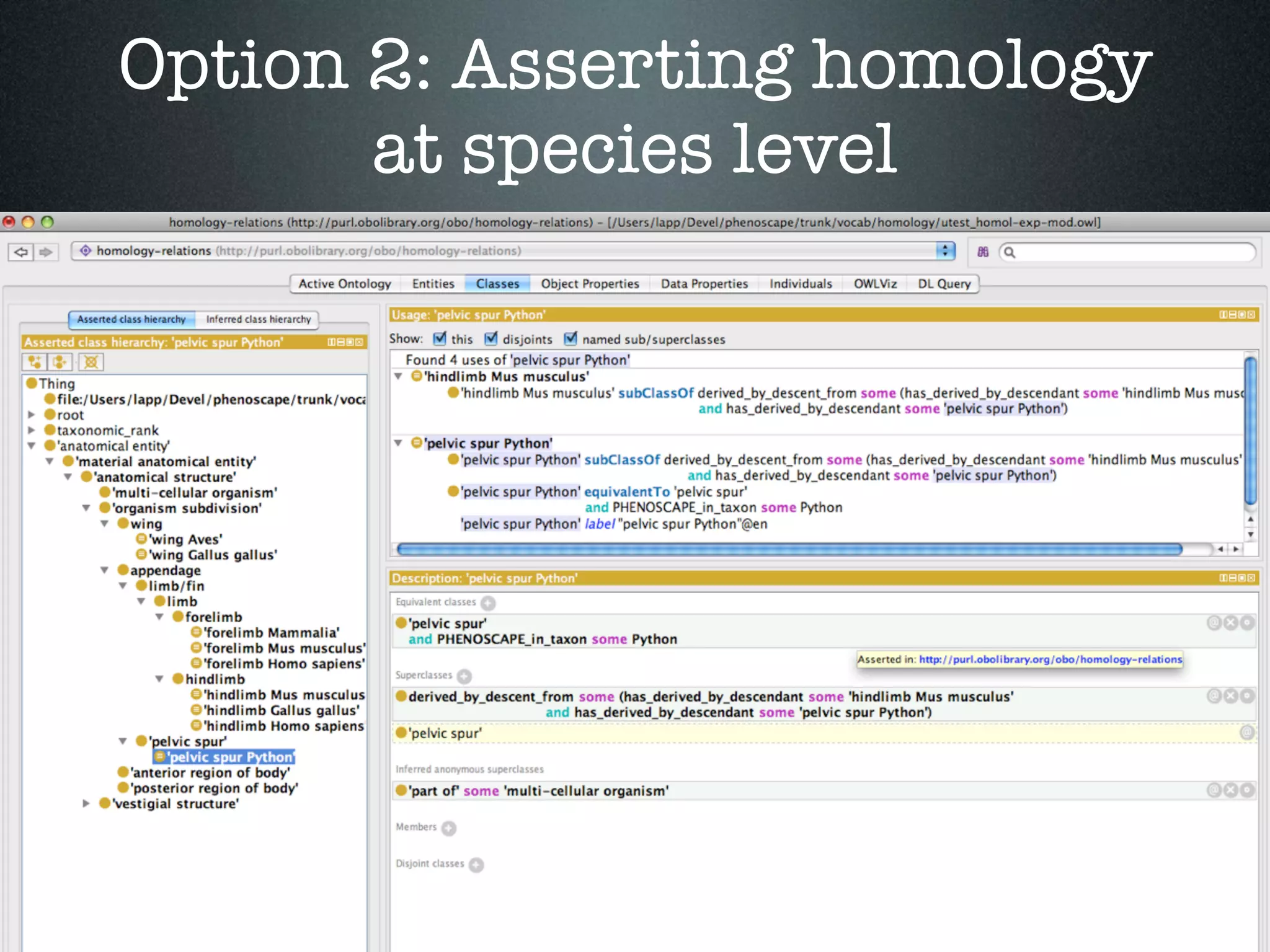
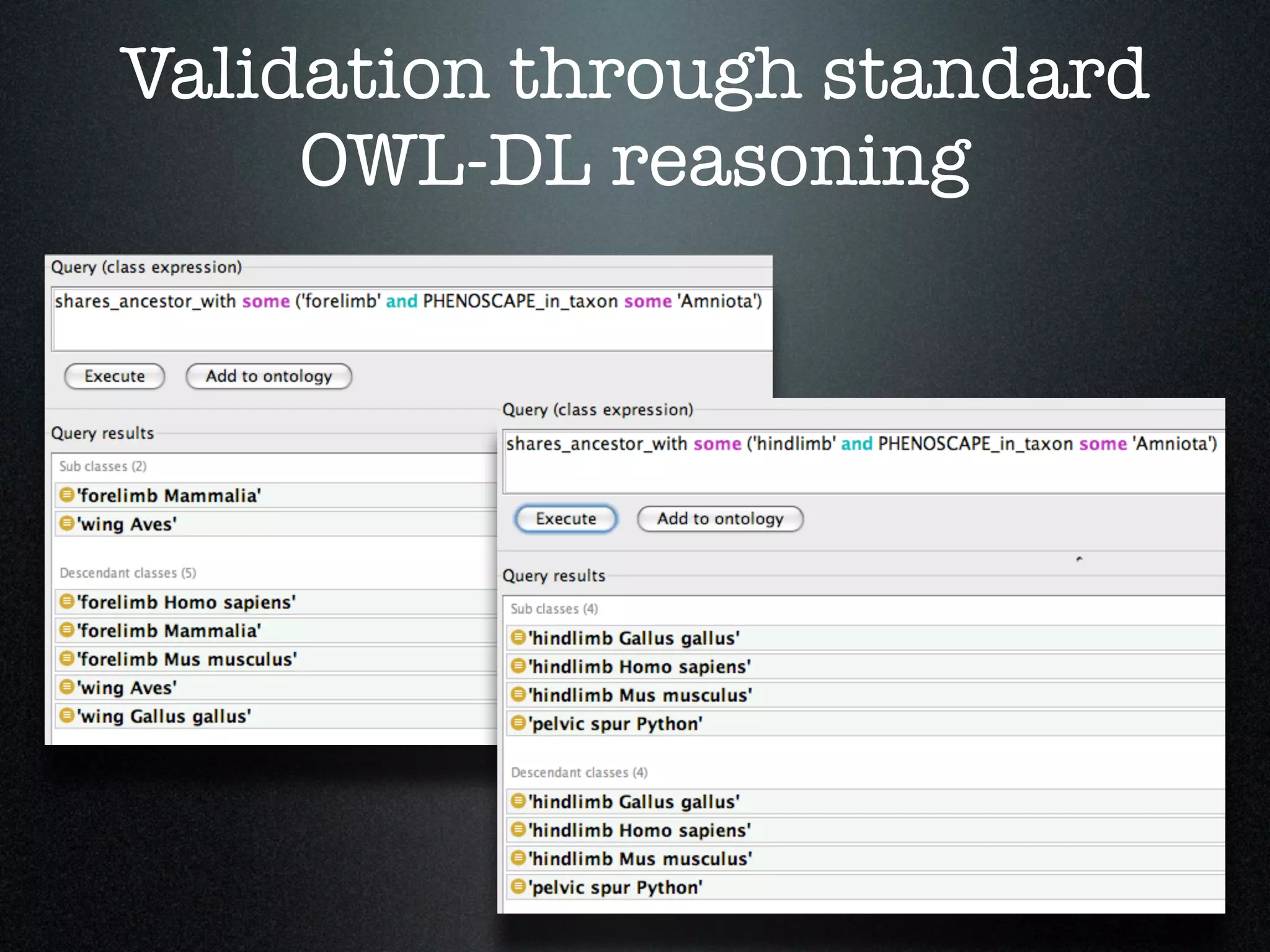
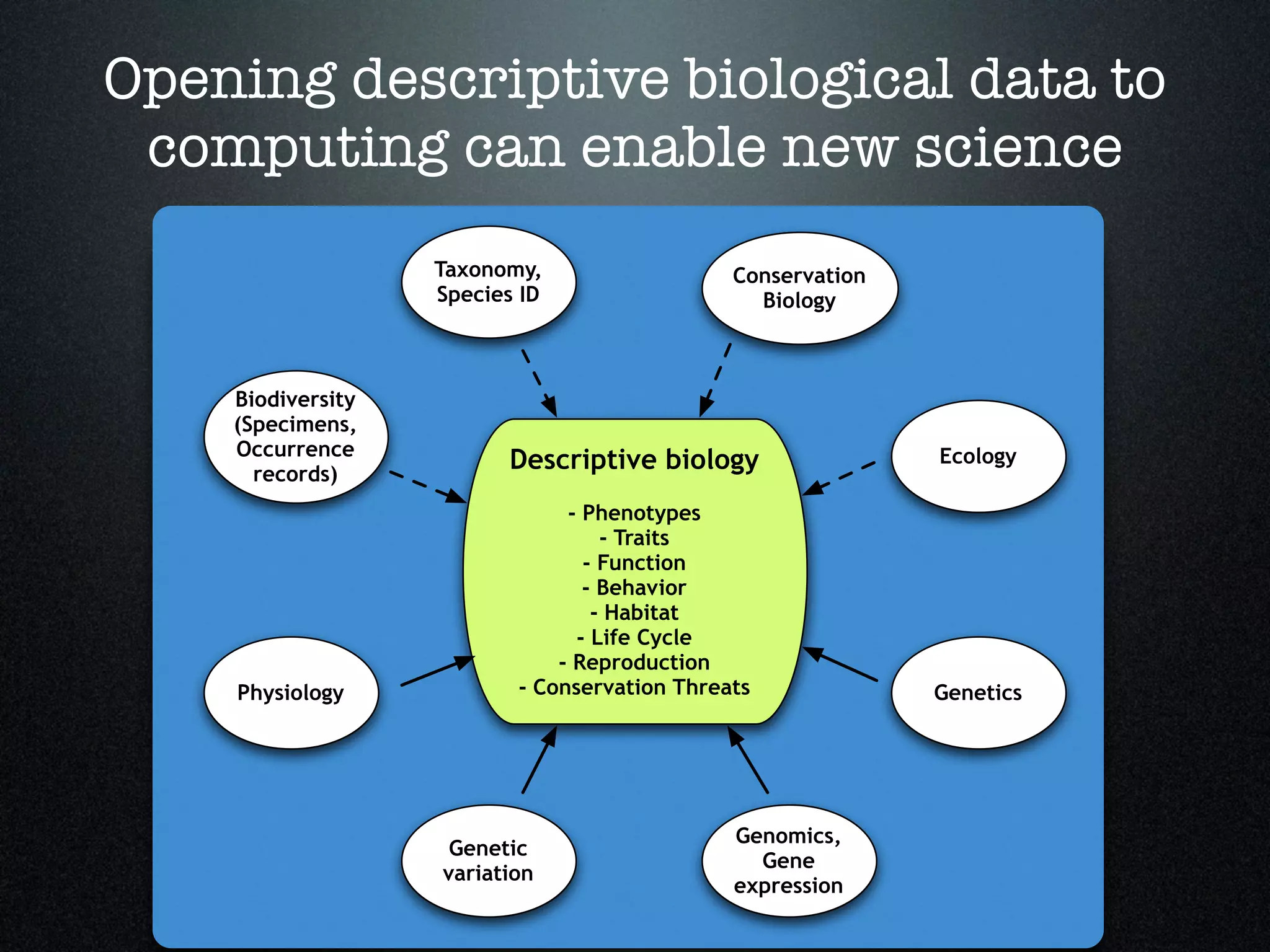
The document discusses the integration of diverse phenotypic data for evolutionary biology, detailing a collaborative project aimed at creating a machine-interpretable database of evolutionary phenotypes. It emphasizes the challenges of computing and encoding phenotypic information and presents various methodologies for data mining, hypothesis generation, and visualizing phenotype variation. The initiative aims to foster synthesis between developmental and evolutionary biology by linking evolutionary characters to genomic data and facilitating research on phenotype changes across species.