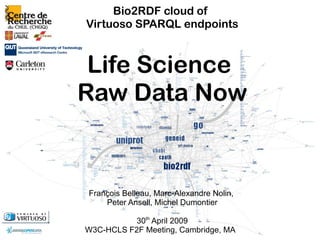
Bio2RDF cloud of Virtuoso SPARQL endpoints
- 1. Bio2RDF cloud of Virtuoso SPARQL endpoints Life Science Raw Data Now François Belleau, Marc-Alexandre Nolin, Peter Ansell, Michel Dumontier 30th April 2009 W3C-HCLS F2F Meeting, Cambridge, MA
- 2. Agenda Why we did Bio2RDF ? ● How we did it ? ● What is know about hexokinase ? ● Where we are going ? ●
- 3. The problem According to NAR 2009 Database collection 1170 public databases exists. How can they be integrated to behave like a global coherent resource ?
- 4. Public map of 1744 namespaces according to BioMoby, NAR, SRS, GO, NCBI, UniProt
- 5. Bio2RDF vision in 2007 Johanne Luciano vision for knowledge integration in 2005 W3C vision of semantic web in 2006
- 6. Bio2RDF Mouse and Human Atlas map in 2008 65 millions triples
- 7. Bio2RDF actual contribution to the Linked Data cloud Linked data cloud in 2007 Linked data cloud in March 2009 http://linkeddata.org/ http://esw.w3.org/topic/TaskForces/CommunityProjects/LinkingOpenData/DataSets/Statistics
- 8. Bio2RDF cloud map of 2,3 billions triples in 2009
- 9. Why do it ? Not to replace HTML or XML by an other new format, RDF and OWL, but to answer science question by submiting SPARQL query over the global knowledge base accessible through the Internet to the Life Science SPARQL endpoints cloud.
- 10. Solution Bio2RDF approach to the data integration problem in bioinformatics : Apply the semantic web approach based on RDF, OWL and SPARQL technologies.
- 11. How we did it ? Bio2RDF architecture
- 12. Our design principles http://www.w3.org/DesignIssues/LinkedData http://bio2rdf.wiki.sourceforge.net/Banff%20Manifesto
- 13. YeastHub design in 2005 Conversion of Dataset to RDF ● Use of Sesame Triplestore ● SeRQL query interface ● http://www.ncbi.nlm.nih.gov/pubmed/15961502
- 14. Bio2RDF at ISMB 2005 the begining Thanks to Kei Cheung, Johanne Luciano, Eric Neumann and Christopher Baker they draw the lines.
- 15. Bio2RDF realtime rdfiser in 2007
- 16. Actual Architecture Offline rdfising process ● ● Virtuoso SPARQL endpoints network ● Namespace resolution through DNS subdomain
- 17. Main REST services Describe a ressource by a dereferencable URI ● http://bio2rdf.org/ns:id ● Global services over federated endpoints ● http://bio2rdf.org/links/ns:id ● http://bio2rdf.org/search/searchedTerm ● Targeted services to a specific endpoint ● http://bio2rdf.org/linksns/ns2/ns1:id ● http://bio2rdf.org/searchns/ns/searchedTerm ● other services are available. ●
- 18. Describe service implementation http://bio2rdf.org/ns:id ● Corresponding SPARQL query : ● CONSTRUCT { ● ?s ?p ?o . } WHERE { ?s ?p ?o . FILTER(?s = <http://bio2rdf.org/ns:id>). } Submited at this URL ● http://ns.bio2rdf.org/sparql?query=... ● Based of DNS subdomain resolution service –
- 19. Bio2RDF JSP server software http://sourceforge.net/projects/bio2rdf/
- 20. Peter Ansell is writing the Bio2RDF JSP server The software transform Bio2RDF URIs to SPARQL ● queries in real time. Its aim is to access normalised RDF information ● located in multiple endpoints using the concept of Public Namespaces and Private Record Identifiers and distributed SPARQL queries which are matched to the content in each endpoint. Each of the following databases have normalisation ● rules which normalise them back to bio2rdf.org URI's :Dbpedia, Drugbank, LinkedCT, HCLS KB/Neurocommons, Diseasome, Dailymed, Bioguid DOI
- 21. Bio2RDF.war package future Provide more pipes to perform integrated actions without ● having to put HTTP SPARQL requests into a workflow system when a URI resolution can perform the query in a distributed and normalised manner more efficiently Bring together the current distributed efforts to provide a ● complete HTML redirection registry so that a large percentage of Bio2RDF namespaces can be redirected with http://bio2rdf.org/html/namespace:identifier Form ontologies describing the query type, provider, rdf ● normalisation rule, namespace paradigm Integrate http://rdf.myexperiment.org/sparql and similar ● workflow RDF endpoints so that scientific workflows can be linked to their data cleanly
- 23. Michel Dumontier will design Bio2RDF.owl ontology next version
- 24. What is known about hexokinase ?
- 25. Submit your query... To the web search engine ● To existing public web site offering data ● integration services; Using Bio2RDF SPARQL endpoints ● Submitting a SPARQL query; ● Using facet browser interface from Virtuoso 6.0 ● server; Dereferencing Bio2RDF search URI; ● Using a Taverna workflow composed of SPARQL ● queries to obtain federated results from KEGG, Entrez Gene and GO;
- 26. The usual unsemantic way
- 27. Existing integrated search services EBI/EB-eye NCBI/Entrez KEGG/DBGET GoPubmed
- 28. By submitting a SPARQL query http://atlas.bio2rdf.org/sparql
- 29. What is know about « hexokinase » with semantic ? select ?t1 ?p2 count(*) where { ?s1 ?p1 ?o1 . FILTER( bif:contains(?o1, quot;hexokinasequot;)) . ?s1 a ?t1 . ?s1 ?p2 ?o2 . } ORDER BY ?t1 ?p2
- 30. Use Virtuoso 6.0 facet browser http://lod.openlinksw.com/
- 32. How can we submit a complex query over the network of SPARQL endpoints ?
- 33. By building a mashup with Taverna 1) Write your complex SPARQL query as if a global graph would be available 2) Identify the needed namespaces and split the query to fetch each data source separetly 3) Build a mashup using a Taverna workflow that instanciate a local triplestore 4) Execute your complex query locally on the mashup
- 34. The SPARQL query needed (dont try this home, do it on the web !)
- 35. Get the list of genes from KEGG pathways of a specified taxon Clear graph ● Get KEGG pathways list for a ● specific taxon For each pathway get genes ● list and import instances Count the number of genes ● found http://www.myexperiment.org/workflows/747
- 36. Insert into local triplestore GeneID genes and KEGG pathways Get the list of genes ● Get the list of pathways ● Insert into local triplestore ● each corresponding graph http://www.myexperiment.org/workflows/748
- 37. Insert into local triplestore the needed GO annotations Get the GO annotations for ● each gene
- 38. Finally, the neeeded query merging KEGG, Entrez Gene and GO together
- 40. Bio2RDF's mirrors http://quebec.bio2rdf.org/ http://qut.bio2rdf.org/
- 42. Life Science Raw Data Now http://quebec.bio2rdf.org/download
- 43. Visit our Wiki rdfiser cookbook http://bio2rdf.wiki.sourceforge.net/
- 44. Bio2RDF news http://bio2rdf.blogspot.com/ http://www.slideshare.net/search/slideshow?q=bio2rdf http://scholar.google.com/scholar?q=bio2rdf http://groups.google.ca/group/bio2rdf
- 45. Our 2009 objectives Get approval from data provider to distribute ● RDF dump and publish SPARQL endpoints (UniProt, BioCyc, Pathway Commons, Bind are in); Start using Virtuoso 6 cluster; ● Design more services accessible with REST ● protocol via our JSP package; Recruit mirror server; ● Develop new rdfiser program in a community ● effort;
- 46. Thanks Jean Morissette, Nicole Tourigny The Bio2RDF community ● Centre de recherche du CHUL ● Université Laval ● Dumontier Lab ● QUT eResearch Center ● Openlink Virtuoso ●
