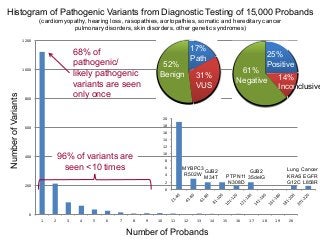
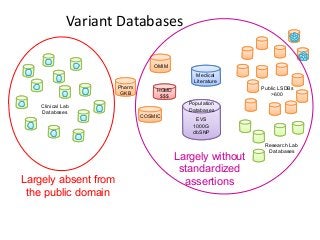
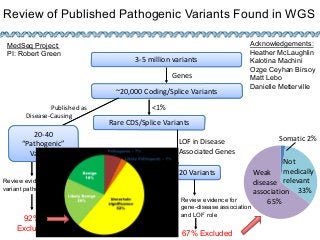
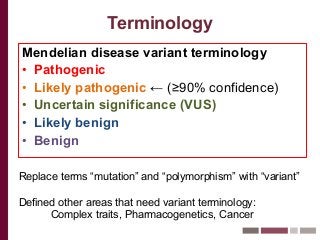
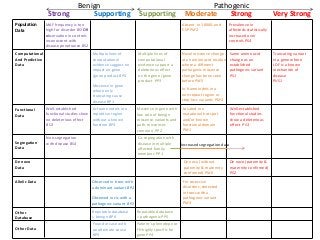
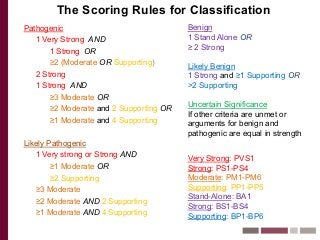
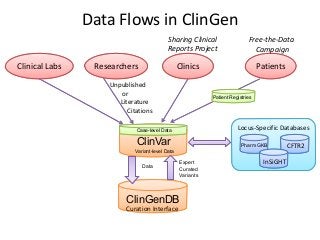
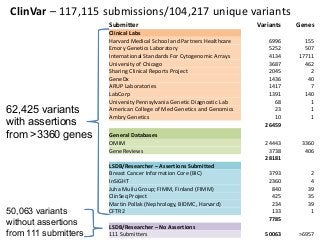
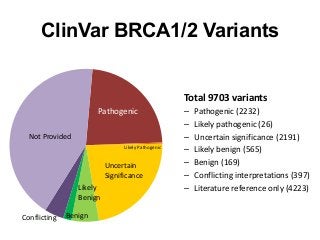
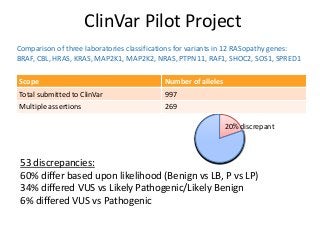
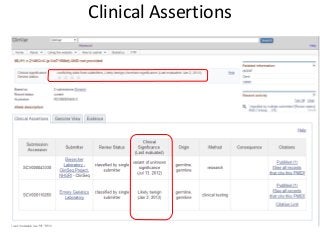
Recently, three NIH-funded efforts were aligned with the National Center for Biotechnology Information’s (NCBI) ClinVar database under the collaborative Clinical Genome Resource Program (ClinGen - http://www.iccg.org/about-the-iccg/clingen). ClinGen is developing interconnected resources for the community to improve our understanding of genomic variation and optimize its use in genomic medicine. A unique aspect of ClinGen is that it represents a strong public-academic-private partnership that relies on the collaboration between NIH, academic and commercial genetic testing laboratories. The project includes the development of standards for variant interpretation as well as data submission and sharing. ClinVar, launched in April 2013, is a cornerstone of the project as it serves as the primary site for deposition and retrieval of variant data and annotations. As of February 1st, 2014 ClinVar contains 73,487 submissions across 18,702 genes (66,956 unique variants) with interpretations from OMIM, GeneReviews, 60 laboratories, and 23 locus-specific databases (LSDBs). The dataset includes 5454 variant submissions (2095 unique variants) from the Sharing Clinical Reports Project (SCRP - http://sharingclinicalreports.org) on BRCA1/2 and 4100 copy number variants from the International Standards for Cytogenomic Arrays consortium. New policies and data structures are being considered to support controlled access to patient-level data. ClinGen is currently working with many laboratories and LSDBs to support robust mechanisms to share their data in an ongoing manner and increase the content of structured data and supporting evidence. Other parts of the project include computational and machine-learning approaches for identifying clinically relevant variants, and the development of expert working groups across many clinical domains to support consensus-driven evidence-based curation of genes-disease associations and genomic variant interpretations. Groups have already been formed in the areas of cardiovascular disease, hereditary cancer, metabolic disease, rasopathies, congenital muscular dystrophy, and developmental delay. The project is also interfacing with a large and diverse community of stakeholders including professional organizations, patient advocacy groups, regulatory agencies, research consortia and other projects from both national and international sites which is facilitated by working with the existing International Collaboration for Clinical Genomics (ICCG - http://www.iccg.org). This talk will give an overview of the ClinGen resource and progress made to date.