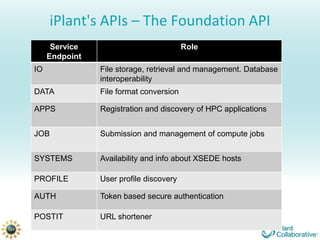
The iPlant Collaborative provides cyberinfrastructure resources for life science research through its data storage system, analysis tools in the Discovery Environment, cloud computing resources on Atmosphere, and other services accessible through portals or APIs to help address challenges of data volume, fragmented tools and lack of reproducibility in biological research. It offers access to petabytes of data and thousands of computational cores through tools for data management, analysis, visualization and sharing of results.