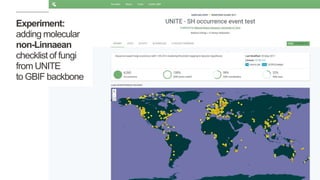
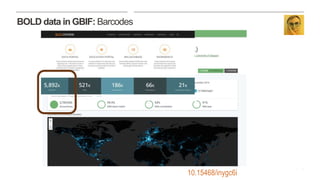
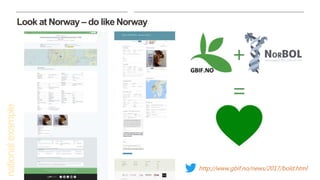
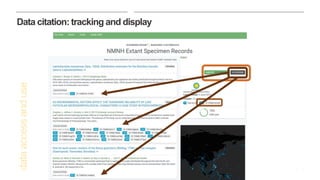
This document discusses GBIF's efforts to bridge biodiversity evidence through data standards by incorporating molecular data like sequences. It describes an experiment to add a non-Linnaean checklist of fungi from the UNITE database to the GBIF backbone taxonomy. This would allow indexing and displaying georeferenced sequence data in the same way as specimen and observation data. Checklists are seen as key, and stable but dynamic digitally citable sources are needed. Successful examples like Norway adding Barcode of Life Data are held up. The document advocates cross-linking all biodiversity data through open sharing on platforms like GBIF and tracking use through data citation.