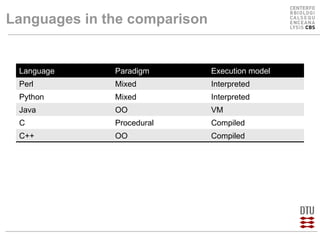
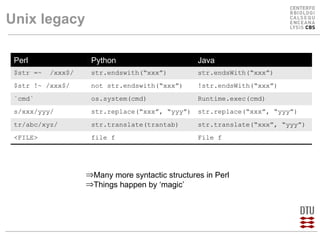
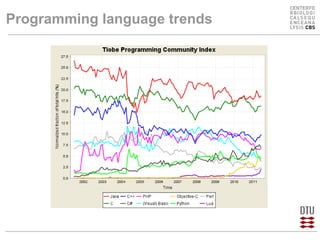
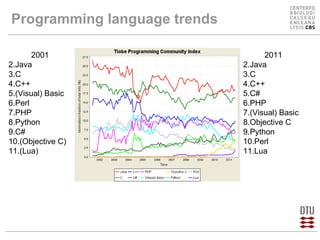
This document compares several programming languages for their suitability for bioinformatics education. It discusses trends in programming language usage over time and compares Perl, Python, Java and C++ in small bioinformatics tasks by measuring lines of code. It also evaluates these languages from a software engineering perspective. Finally, it reviews major bioinformatics libraries like BioPerl, BioPython and BioJava and their commit activity over time. The document concludes that scripting languages are better suited for teaching bioinformatics and that new languages and libraries make teaching the subject easier.



![Software engineering perspective Effort (/time) to solve a problem is proportional to the number of lines needed for the solution [1] The number of bugs per line of code is constant regardless of the language used [2] [1] F Brooks 1995 [2] L Hatton 1995](https://image.slidesharecdn.com/programminglanguagesvienna-110719155403-phpapp01/85/Programming-languages-vienna-8-320.jpg)