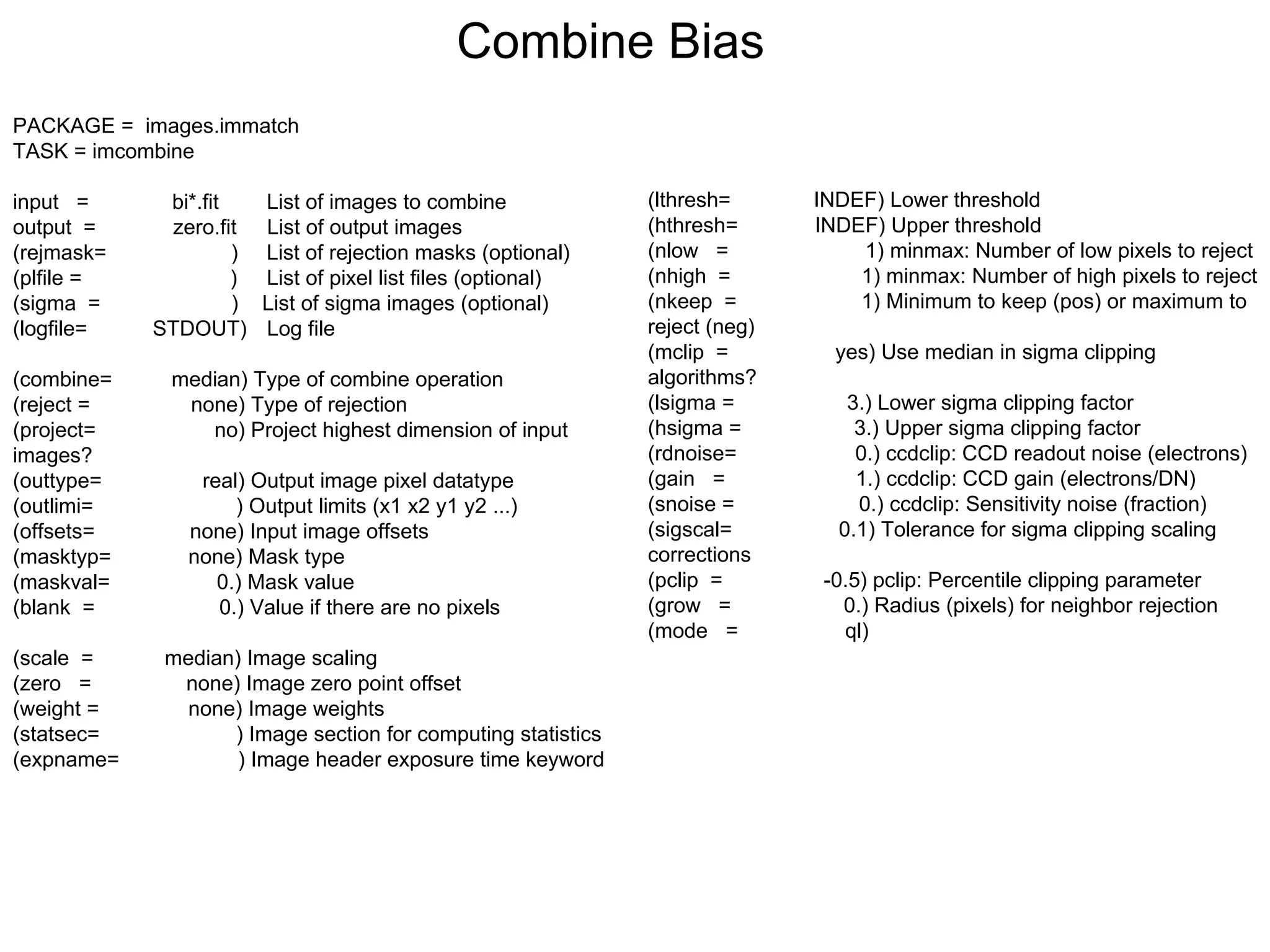
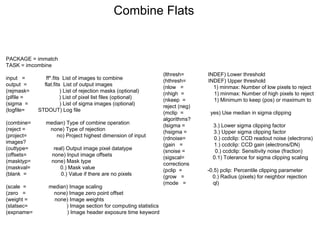
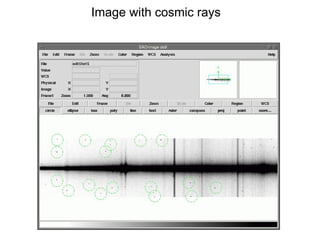
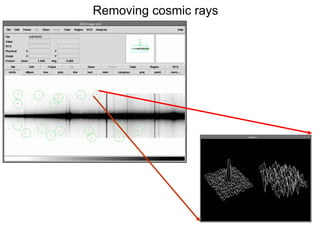
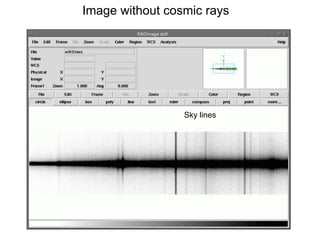
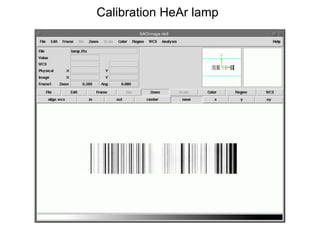
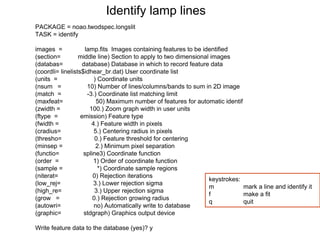
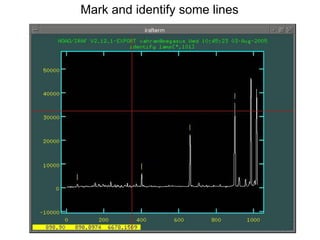
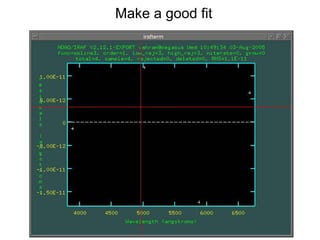
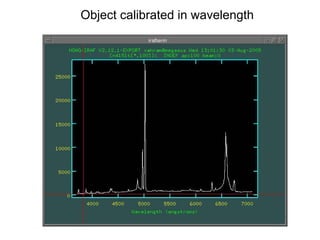
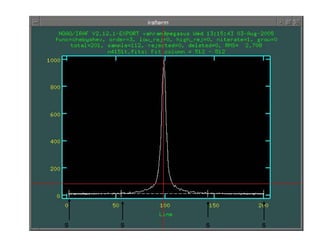
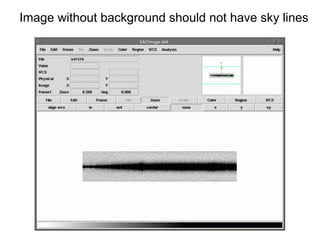
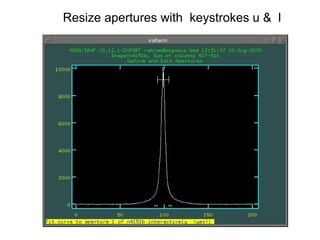
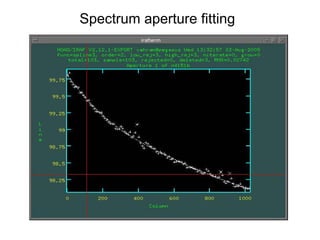
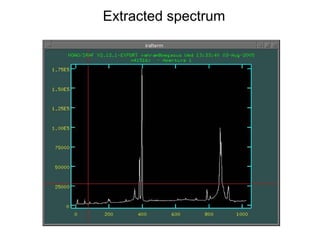
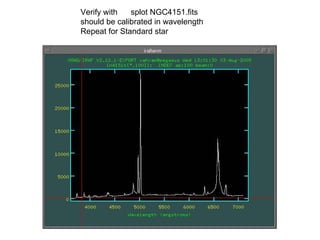
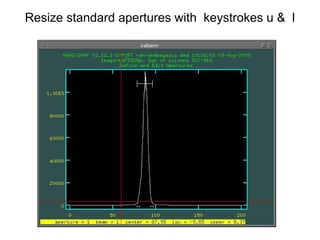
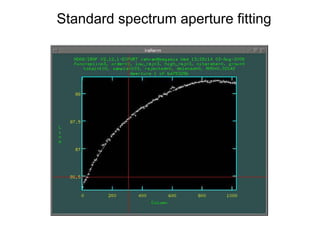
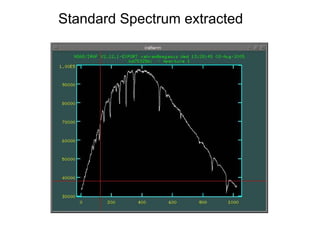
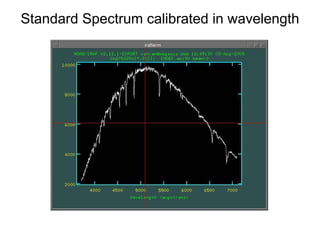
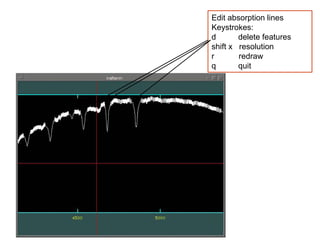
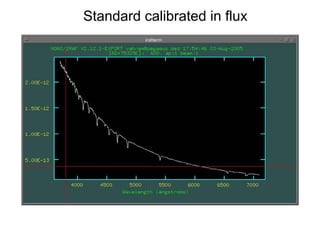
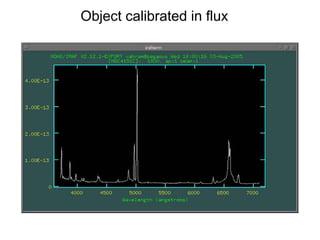
The document provides instructions for combining bias frames, creating a master flat field, removing the flat field from science images, detecting and removing cosmic rays, identifying emission lines in a HeAr lamp calibration frame, and adding header information to processed science and standard star images.