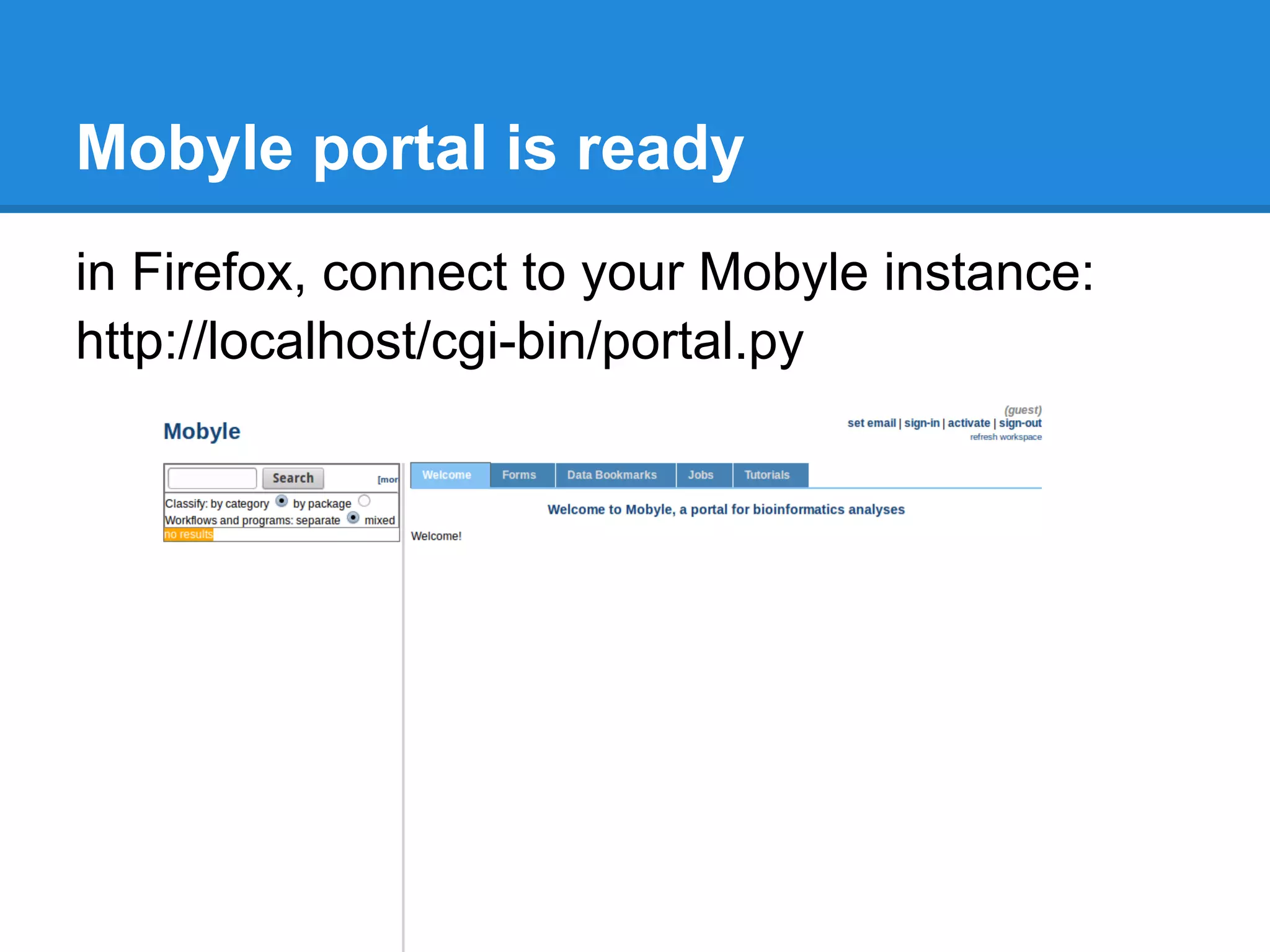
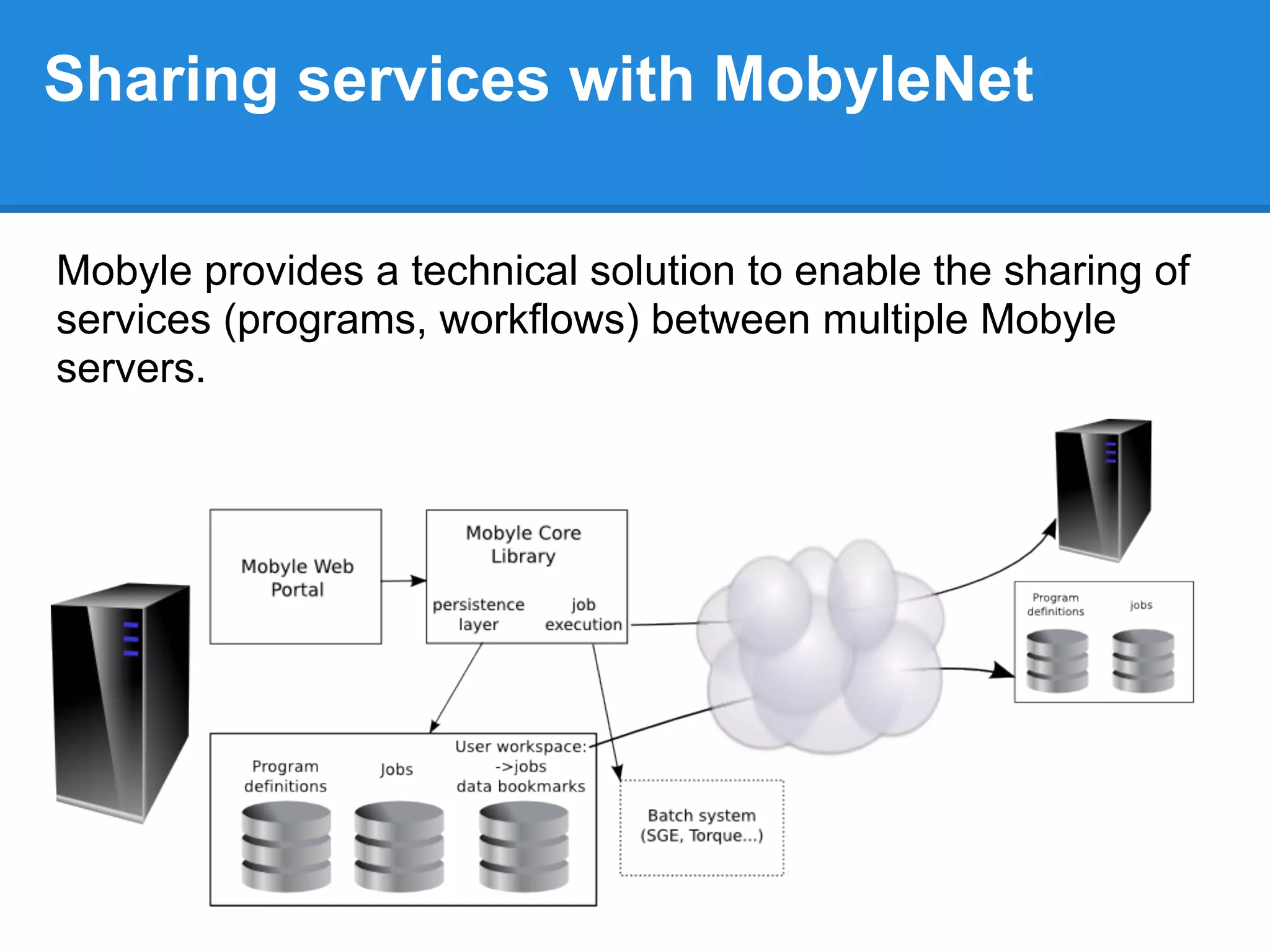
The document outlines the installation and configuration procedures for Mobyle, a platform for bioinformatics services, covering aspects such as architecture, service deployment, and integration with execution systems. Key requirements and configuration details for Apache, Python, and necessary libraries are provided along with instructions for managing services and controlling access. It also highlights the use of virtual machines and specific file paths for various tasks related to Mobyle operation and service management.









![setup.cfg: a way to automate installation
[install]
install_core=/opt/mobyle
install_htdocs=/var/www/mobyle/htdocs
install_cgis=/var/www/mobyle/cgis
install_bmid=True
install_bmps=True
sudo python setup.py install](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-14-2048.jpg)
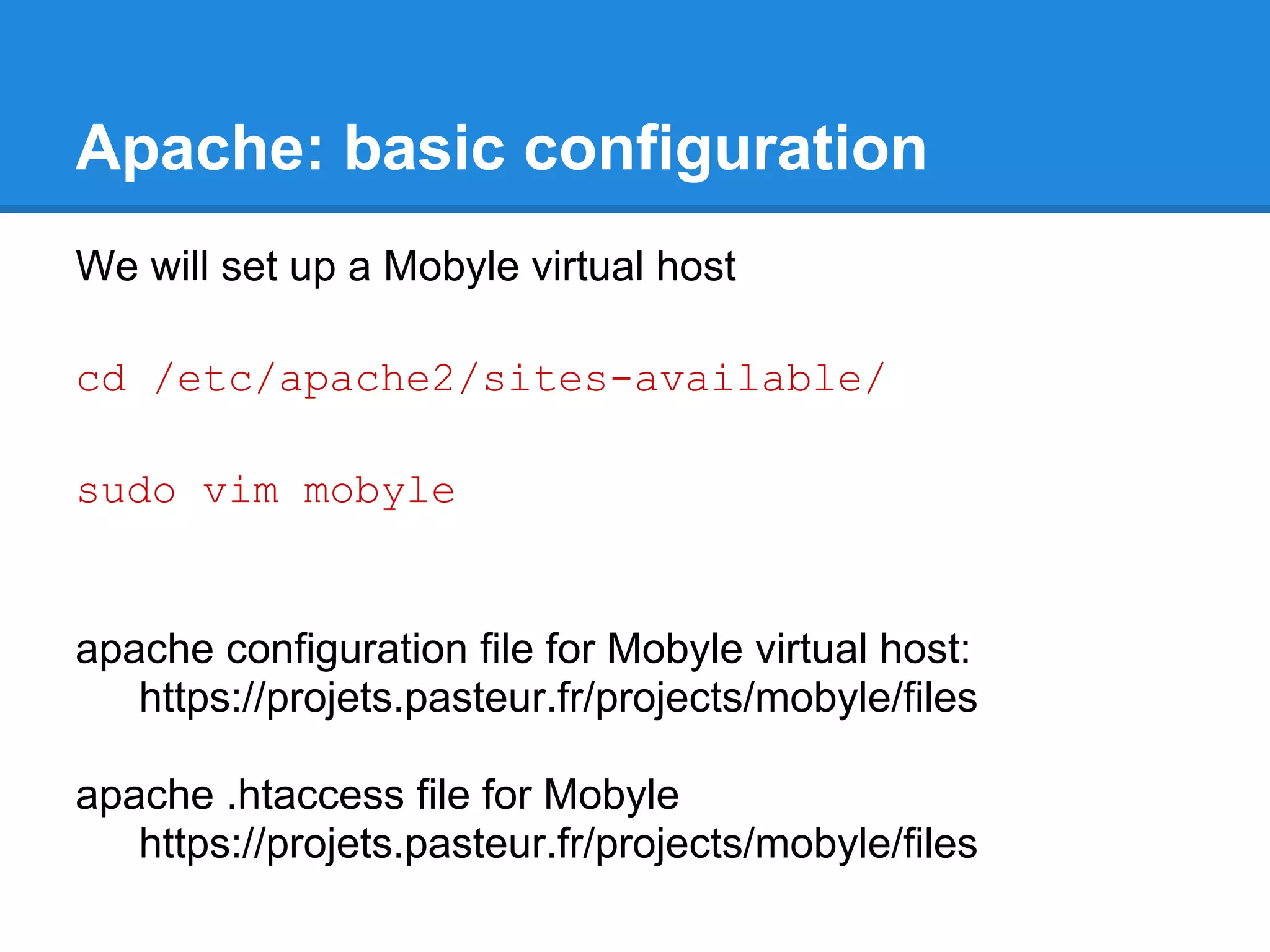
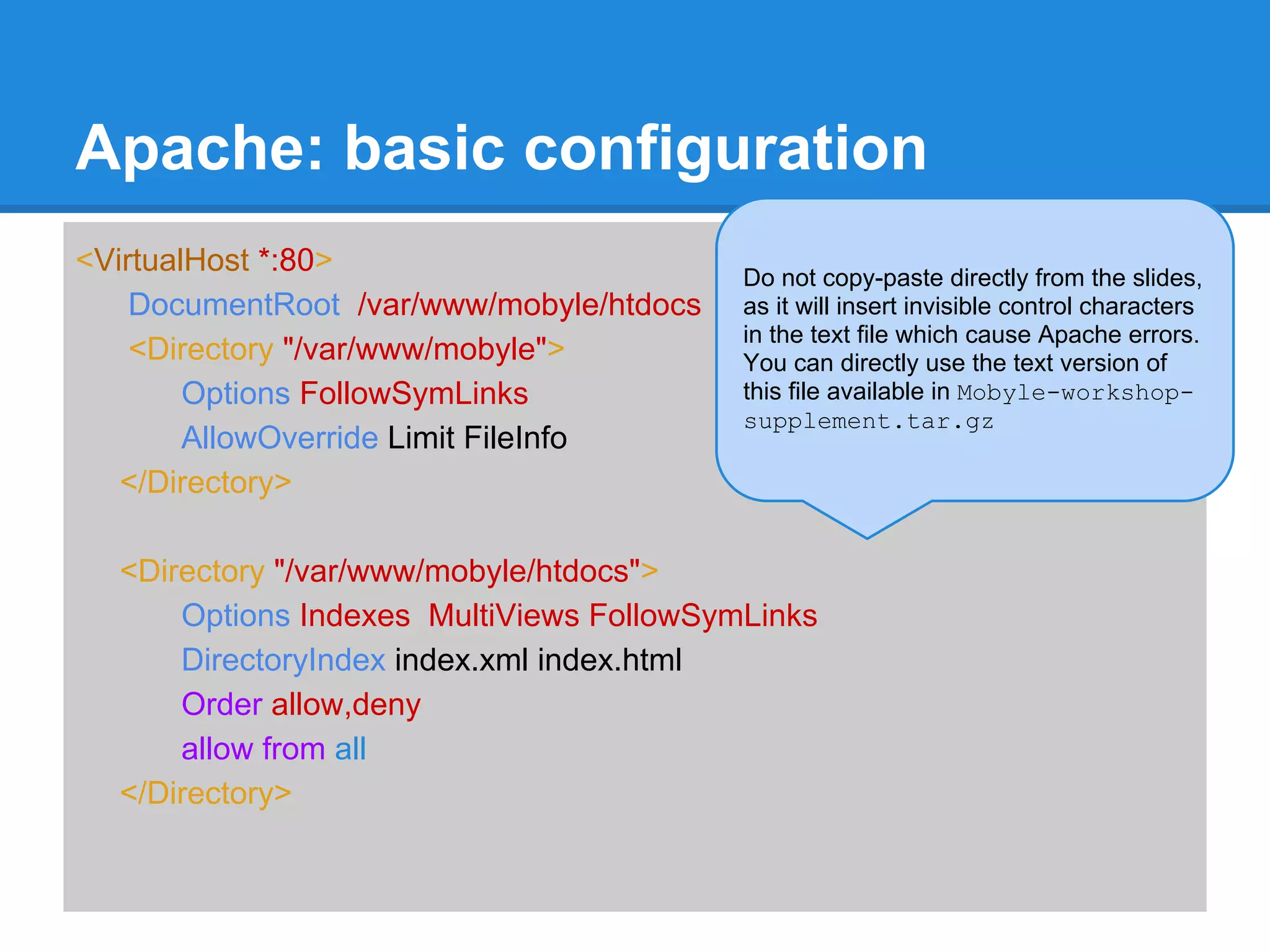
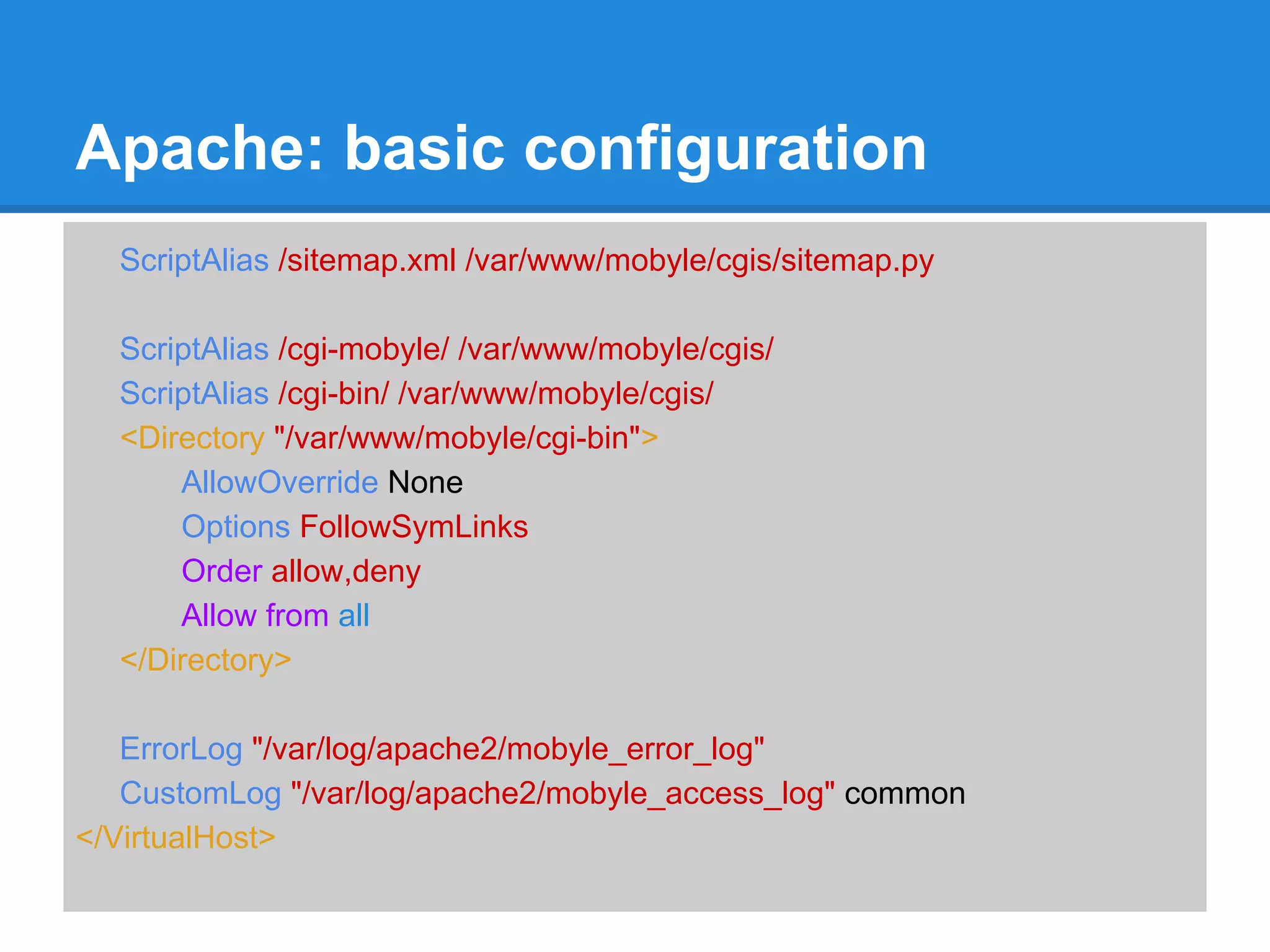
![Mobyle: basic configuration
cd /opt/mobyle
sudo cp Example/Local/Config/Config.
template.py Local/Config/Config.py
sudo vim Local/Config/Config.py
ROOT_URL = "http://localhost/"
HTDOCS_PREFIX= ""
CGI_PREFIX= "cgi-bin"
MAINTAINER= [""]
HELP= [""]
MAILHOST= "localhost"](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-21-2048.jpg)
![Apache: advanced config & security
Do not copy-paste directly from the slides,
RewriteEngine on as it will insert invisible control characters
#Do not show hidden files content in the text file which cause Apache errors.
You can directly use the text version of
RewriteCond %{REQUEST_URI} /. [OR] this file (htaccess) available in Mobyle-
RewriteCond %{REQUEST_URI} /ADMINDIR workshop-supplement.tar.gz and
copy it as .htaccess in Mobyle
RewriteRule .* - [F,L] htdocs folder
#allow saving results
RewriteCond %{REQUEST_URI} ^/data/jobs(.*)
RewriteCond %{QUERY_STRING} ^save$
RewriteRule (.*)/([^/]+)$ $1/$2 [E=SAVEDFILENAME:$2]
Header set Content-Disposition "attachment; filename="%{SAVEDFILENAME}
e"" env=SAVEDFILENAME](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-22-2048.jpg)
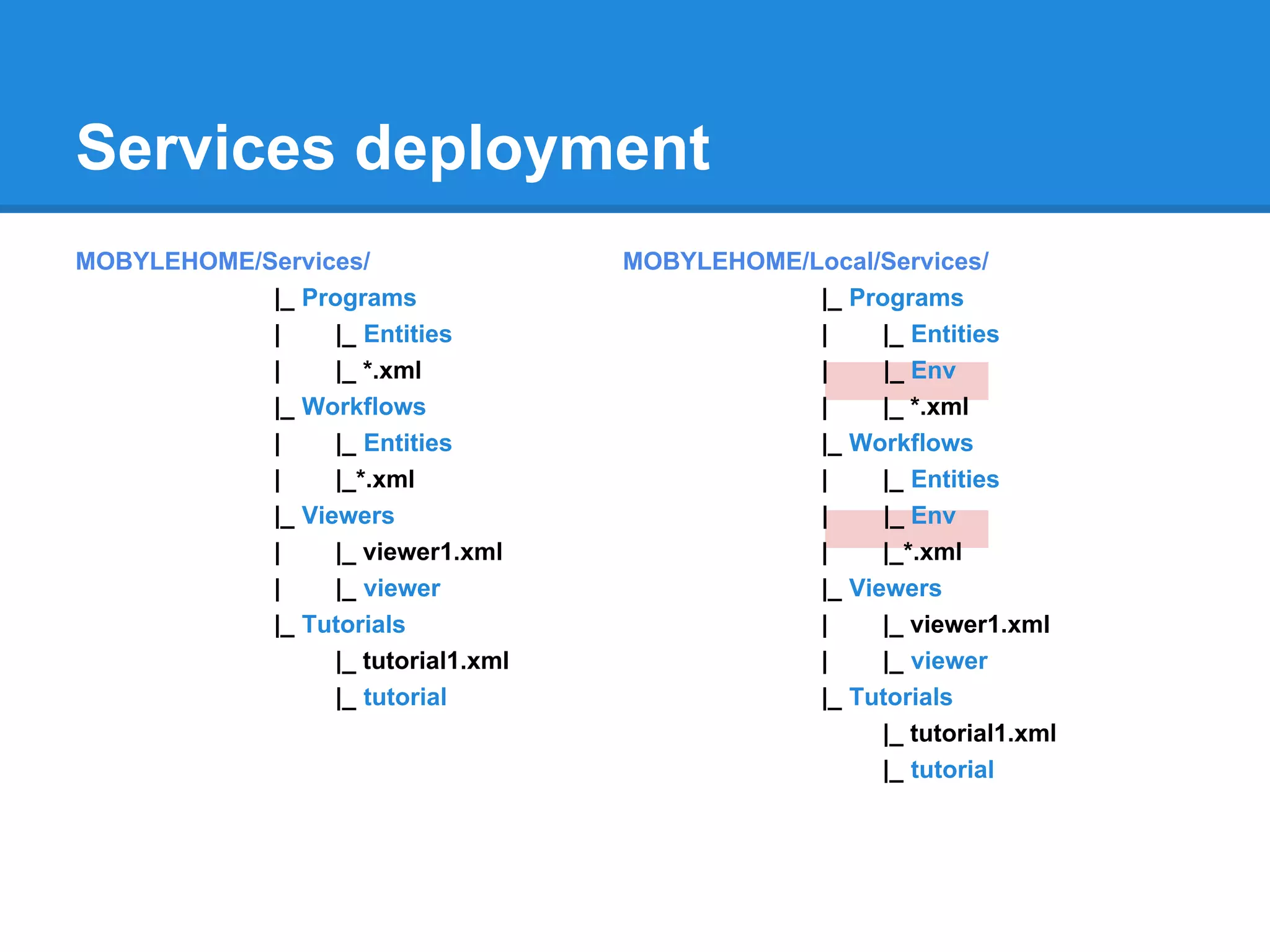
![How to deploy
Mobyle configuration:
LOCAL_DEPLOY_INCLUDE = { 'programs' : [ '*' ] ,
'workflows': [ '*' ] ,
'viewers' : [ '*' ] ,
'tutorials' : [ '*' ] ,
}
LOCAL_DEPLOY_EXCLUDE = { 'programs' : [ '' ] ,
'workflows': [ '' ] ,
'viewers' : [ '' ] ,
'tutorials' : [ '' ] ,
}
tool deployment:
mobdeploy -s local -p all deploy mobdeploy -s local -p prog1,prog2 deploy
mobdeploy deploy mobdeploy -s local -t all deploy](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-26-2048.jpg)
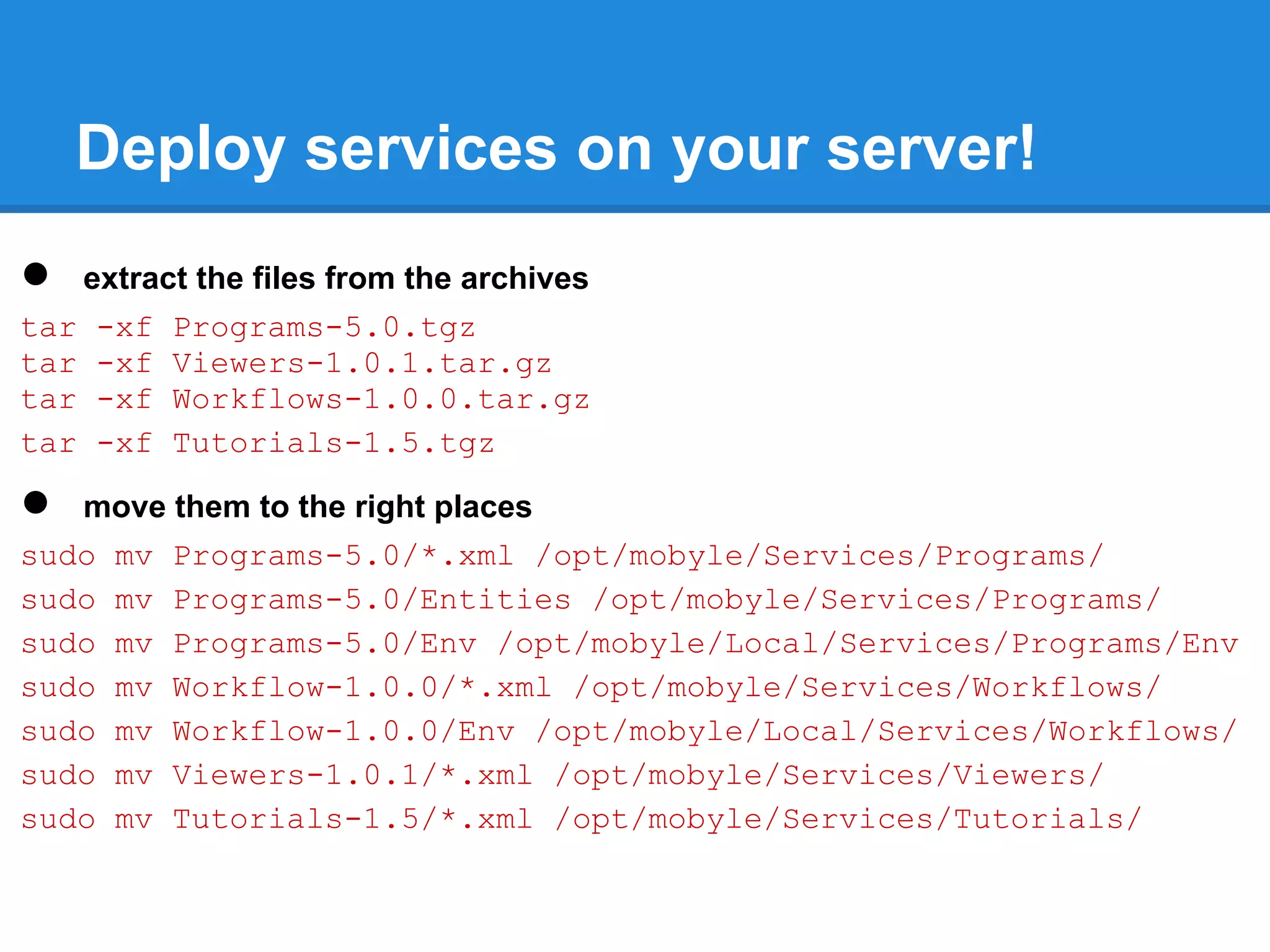
![Deploy services on your server!
● configure deployment
sudo vim /opt/mobyle/Local/Config/Config.py
LOCAL_DEPLOY_INCLUDE = { 'programs' : [ '*' ] ,
'workflows': [ '*' ] ,
'viewers' : [ '*' ] ,
}
LOCAL_DEPLOY_EXCLUDE = { 'programs' : [ 'mafft' ] ,
'workflows': [ '' ] ,
'viewers' : [ '' ] ,
}
● deploy
sudo -u www-data /opt/mobyle/Tools/mobdeploy deploy](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-29-2048.jpg)

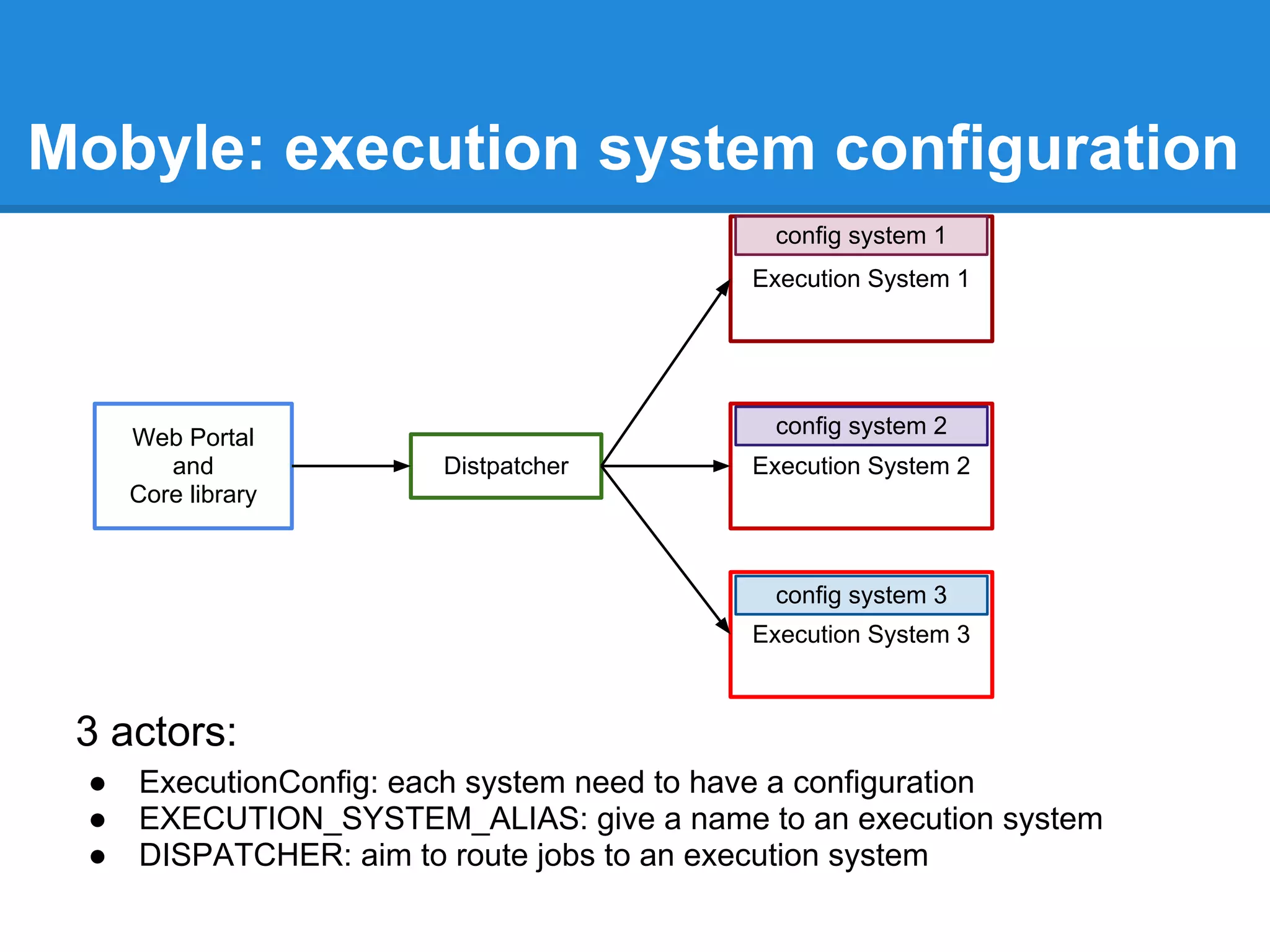
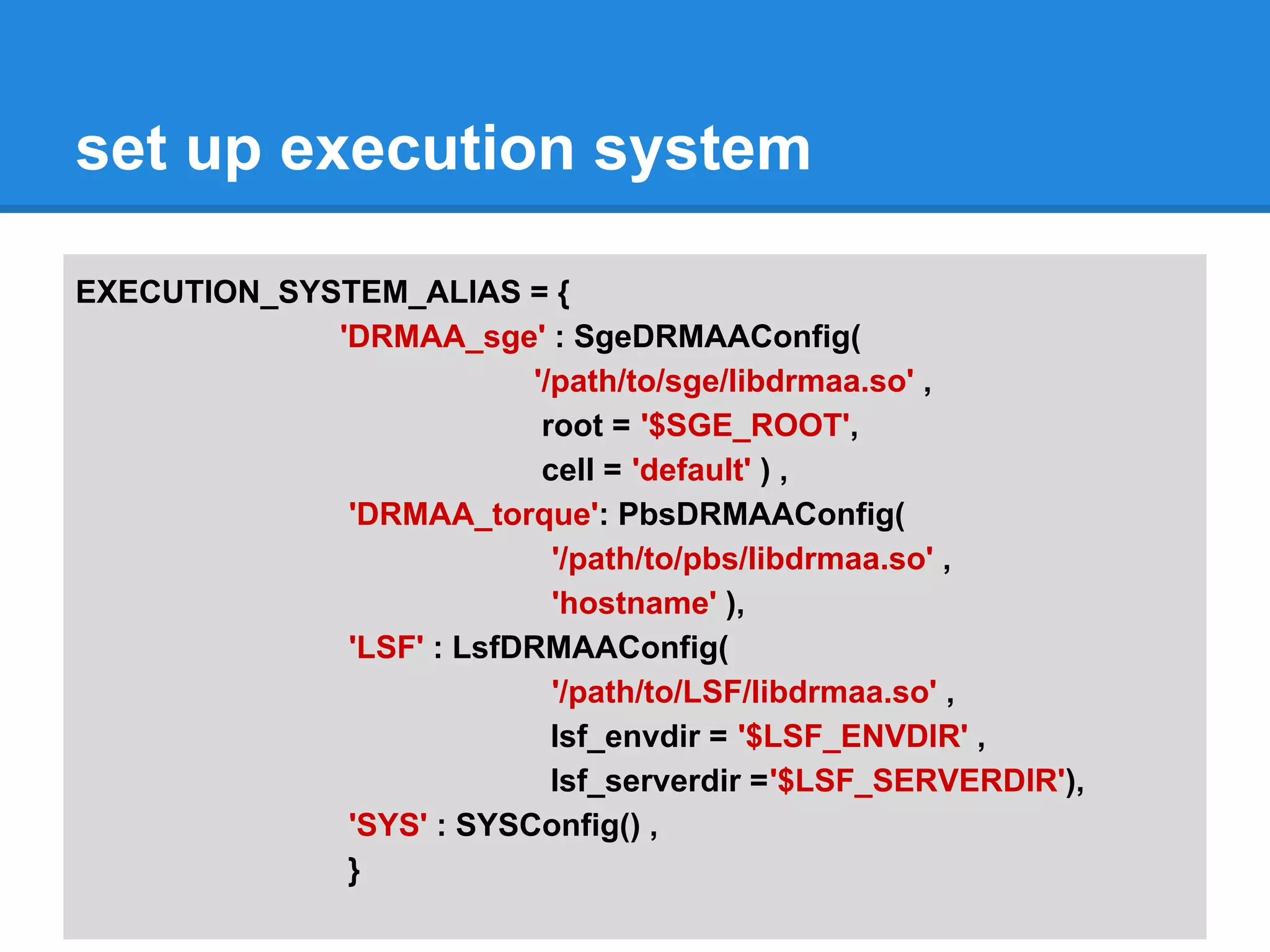
![set up execution system
DISPATCHER = DefaultDispatcher( {
'clustalw' : ( EXECUTION_SYSTEM_ALIAS[ 'DRMAA_sge' ] , 'mobyle' ),
'clustalo' : ( EXECUTION_SYSTEM_ALIAS[ 'DRMAA_torque' ] , 'mobyle' ),
'toppred' : ( EXECUTION_SYSTEM_ALIAS[ 'DRMAA_torque' ] , 'short' ),
'blast2': ( EXECUTION_SYSTEM_ALIAS[ 'DRMAA_sge' ] , 'long' )
'DEFAULT' : ( EXECUTION_SYSTEM_ALIAS[ 'SYS' ] , '' )
})
For the workshop we won't use a cluster, so we will execute all jobs on local
system.
DISPATCHER = DefaultDispatcher( {
'DEFAULT' : ( EXECUTION_SYSTEM_ALIAS[ 'SYS' ] , '' )
})](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-34-2048.jpg)
![binary path
In Mobyle configuration we can add some path to the general web server PATH.
BINARY_PATH = [ "/usr/bin", "usr/local/bin", "/local/Bioinfo/bin" ]
instead of tag env in programs/workflows descriptions,
● this new path is available for all services,
● these locations are added to the PATH (do not replace it), their order is
preserved.
In debian/ubuntu the phylip package binaries are installed in "/usr/lib/phylip/bin"
and all binaries are accessible through the phylip wrapper, e.g. protdist -> phylip
protdist
so we must either:
● modify each interfaces belonging to Phylip package (interfaces)
● add this specific path in our Mobyle PATH](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-35-2048.jpg)
![Set BINARY_PATH on your server
To run correctly PHYLIP programs on your server:
sudo vim /opt/mobyle/Local/Config/Config.py
BINARY_PATH = ["/usr/local/bin", "/usr/lib/phylip/bin"]](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-36-2048.jpg)


![Control access to services
Disable/enable all the portal:
#DISABLE_ALL = False
DISABLE_ALL = True
The list of services is empty
The job submission is disabled
Disable a service:
DISABLED_SERVICES = [ 'local.blast2', 'clustalw*', 'genouest.blast2' ]
Disable a portal:
DISABLED_SERVICES =[ 'genouest.*']](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-39-2048.jpg)
![Control access to services
Restrict the access to one programs or workflows to one
IP or a set of IP
AUTHORIZED_SERVICES = {
'http://mobyle.pasteur.fr/data/services/servers/local/programs/netNglyc.xml' : [ '157.99.*.*' ]
}
○ The service will appear only for the users in the subnet
○ Only the users in the subnet can submit this job](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-40-2048.jpg)
![Control access to services
The last way to control the access is to black list a user
email or an ip.
The black_list.py is located in MOBYLEHOME/Local
users = [ 'blub@web.de', 'kt7mail@gmail.com', 'caca@crotte.fr',
'*@aaa*.com' , 'toto@*', 'titi@*', '*@bidule.fr',
]
host = [ '142.161.25.184', '141.161.25.102']
a message will appear to the user:
you have abused our service. Your are not allowed to run on this server for now.
For more informations contact mobyle@pasteur.fr".
This message is customizable in function emailUserMessage
located in Local/Policy.py file](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-41-2048.jpg)
![Data helpers: Bank configuration
'embl':{
'dataType' : 'Sequence' ,
'bioTypes' : ['Nucleic','DNA'] ,
'label' : 'EMBL Nucleotide Sequence Database',
'command' : ['/usr/local/bin/golden', '%(db)s:%(id)s']
},
'genbank':{
'dataType' : 'Sequence',
'bioTypes' : ['Nucleic','DNA'],
'label' : 'Genbank NIH DNA sequence database',
'command': ['/usr/bin/seqret', '%(db)s:%(id)s -osformat2 fasta -auto -stdout']
},
'fasta':{
'dataType':'Sequence',
'label': 'GenOuest Data Banks',
'command': [ "/opt/mongo/mongo.pl", "%(id)s" ]
}
}](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-42-2048.jpg)
![Data helpers: Bank configuration
sudo vim /opt/mobyle/Local/Config/Config.py
DATABANKS_CONFIG = {
'imgt':{
'dataType':'Sequence',
'bioTypes':['DNA'],
'label': 'IMGT',
'command': ['golden', '%(db)s:%(id)s']
},
'uniprot_sprot':{
'dataType':'Sequence',
'bioTypes':['Protein'],
'label': 'SWISSPROT',
'command': ['golden', '%(db)s:%(id)s']
}}](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-43-2048.jpg)
![Data helpers: format detector/convertor
sudo vim /opt/mobyle/Local/Config/Config.py
DATA_CONVERTER={
'Sequence': [ squizz_sequence('/usr/bin/squizz') ] ,
'Alignment': [ squizz_alignment('/usr/bin/squizz')]
}](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-44-2048.jpg)




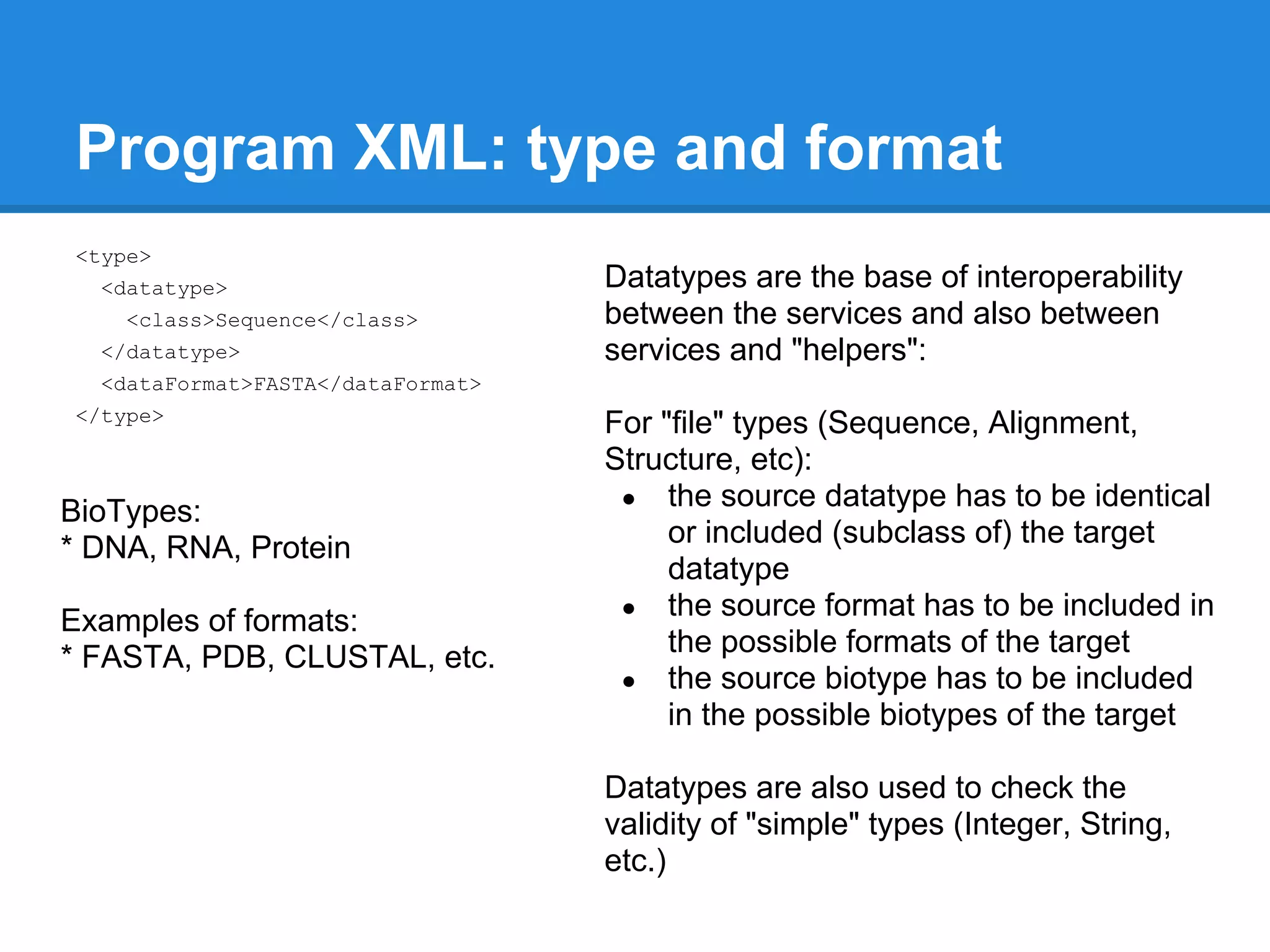
![Program XML: another "simple"
parameter
<parameter>
<name>maxiters</name>
<prompt lang="en">Maximum number of iterations (-maxiters)</prompt>
<type><datatype><class>Integer</class></datatype></type>
<vdef><value>16</value></vdef>
<format>
<code proglang="perl">(defined $value and $value != $vdef) ? " -
maxiters $value" : ""</code>
<code proglang="python">( "" , " -maxiters " + str( value ) )[ value
is not None and value != vdef]</code>
</format>
<comment>
<text lang="en">You can control the number of iterations that MUSCLE
does by specifying the -maxiters option.</text>
[...]
</comment>
</parameter>](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-53-2048.jpg)
![Program XML: an output parameter
<parameter isstdout="1">
<name>muscleHtmlout</name>
<prompt lang="en">Alignment</prompt>
<type>
<datatype>
<class>MuscleHtmlAlignment</class>
<superclass>AbstractText</superclass>
</datatype>
</type>
<precond>
<code proglang="perl">$outformat == 'html' </code>
<code proglang="python">outformat == 'html'</code>
</precond>
<filenames>
<code proglang="perl">(defined $outfile) ? "$outfile" : "muscle.out"</code>
<code proglang="python">( outfile , "muscle.out")[outfile is None]</code>
</filenames>
</parameter>](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-54-2048.jpg)
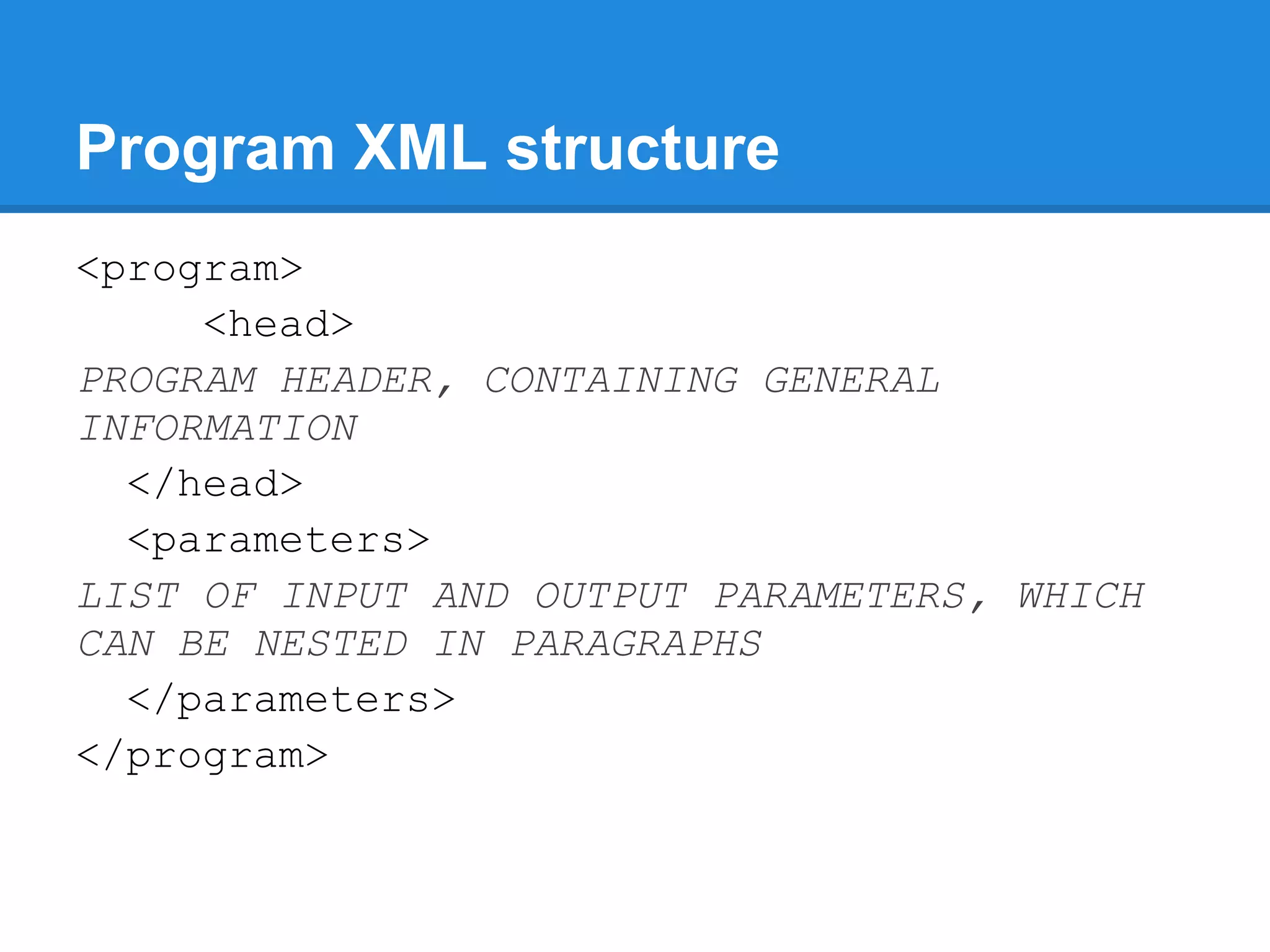
![Program XML: paragraphs
<paragraph>
<name>inputs</name>
<prompt lang="en">Inputs options</prompt>
<parameters>
<parameter issimple="1" ismandatory="1">
<name>sequence</name>
[...]
</parameter>
<parameter>
<name>seqtype</name>
[...]
</parameter>
</parameters>
</paragraph>](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-55-2048.jpg)





![Import/Export Mobyle services with
MobyleNet
PORTAL_NAME = "moi_meme"
Defines the information sent to
PORTALS={ remote execution portals to identify
yourself
'ami_1':{
'url': 'http://ami1.fr/cgi-bin/',
'help' : 'support@ami1.fr',
'repository': 'http://ami1.fr/',
Defines the list of remote services
'services': { 'programs' :[
'golden','dnapars','boxshade','protpars' ], that can be run from your portal
'workflows':[ 'workflow_phylogeny' ]
}
},
'ami_2':{
'url':'http://ami2.fr/mobyle/cgi-bin/',
'help' : 'support@ami2.fr',
'repository' : 'http://ami2.fr/mobyle',
'services': {'programs':['protpars']
}
}
} Defines the list of "your" which you
allow to be run from remote Mobyle
EXPORTED_SERVICES = [ 'abiview','toppred' ] servers](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-64-2048.jpg)
![Import/Export Mobyle services with
MobyleNet
sudo vim /opt/mobyle/Local/Config/Config.py
PORTAL_NAME = "training_session_MYNAME"
PORTALS={
'pasteur':{
'url':'http://mobyle.pasteur.fr/cgi-bin/',
'help':'mobyle@pasteur.fr',
'repository':'http://mobyle.pasteur.fr/',
'services': {'programs': ['protpars']}
}
}
sudo -u www-data /opt/mobyle/Tools/mobdeploy deploy](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-65-2048.jpg)

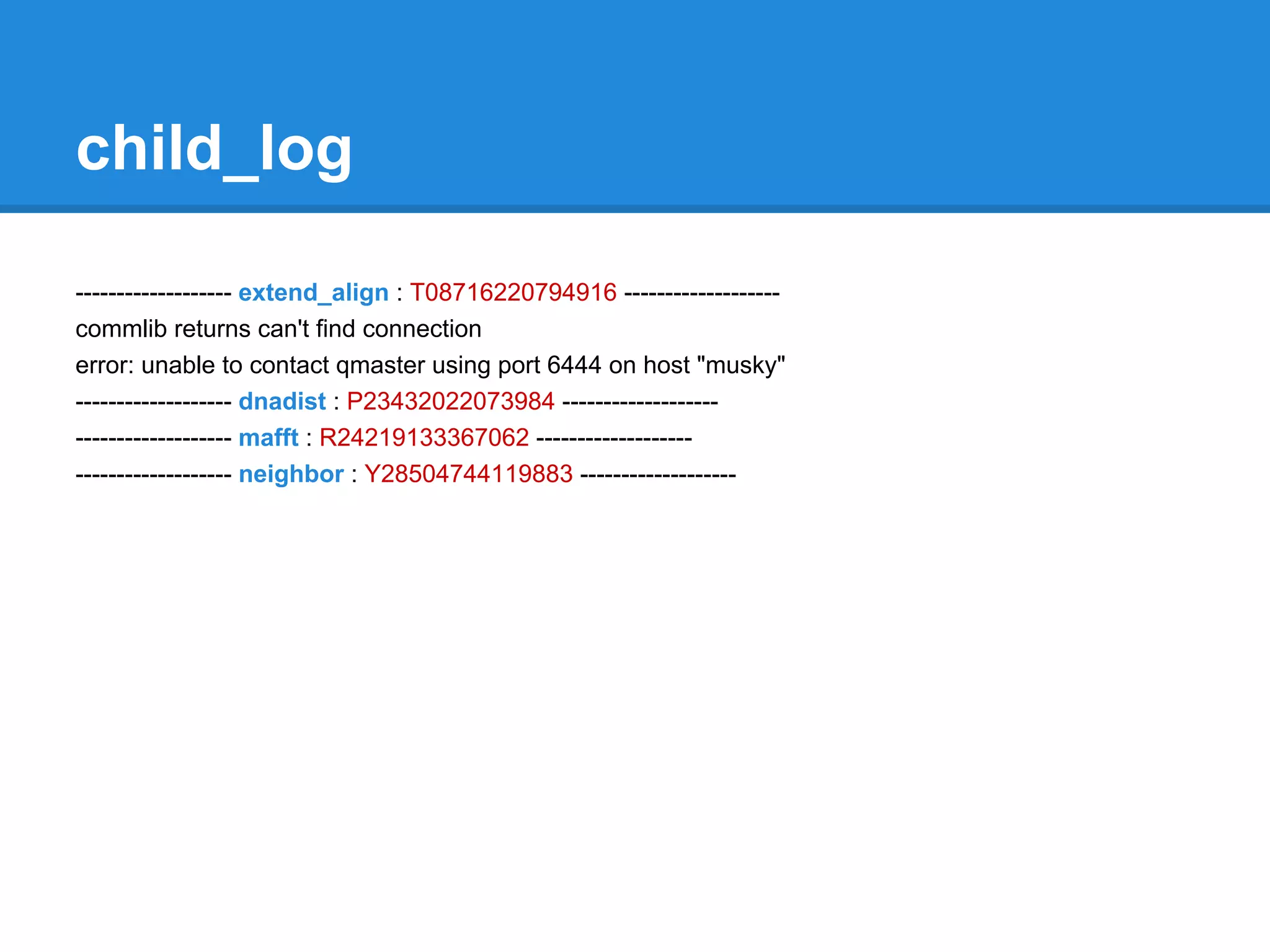
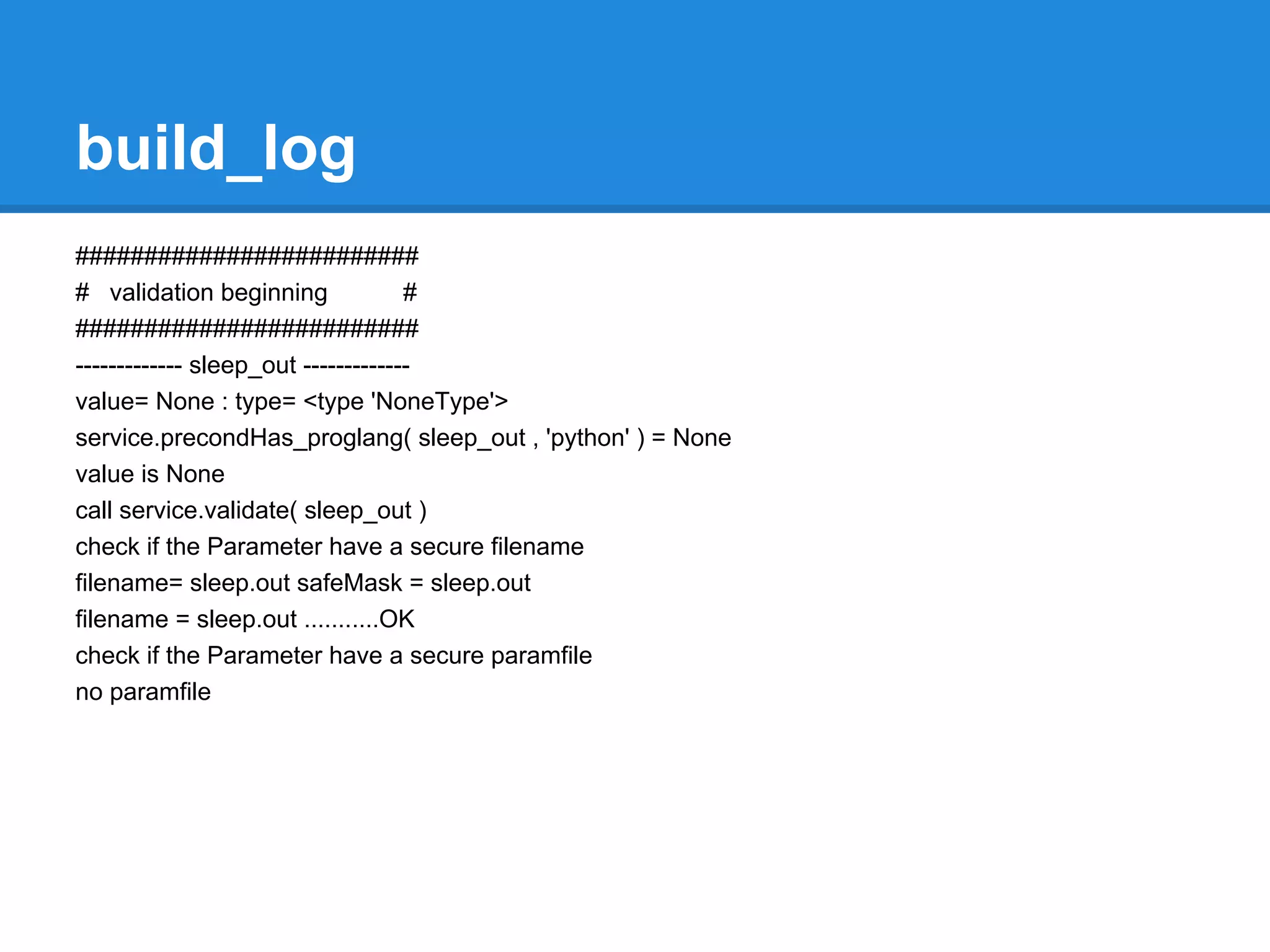
![error_log
Mobyle.MobyleJob : WARNING : MobyleJob.py: L 717 : Fri, 30 Mar 2012 10:57:56 : Potential
Collision: ['/htdocs/data/jobs/mafft/J12393850061893/mafft.out'] input files match the "result" output
parameter mask /htdocs/data/jobs/mafft/J12393850061893/mafft.out
Mobyle.Session.AnonymousSession : ERROR : Session.py: L 491 : Thu, 29 Mar 2012 14:06:57 :
session/J05040996510983 : the data af50ab3b273d6f3bd97b396ed3771bc7.data ( 3388999 ) cannot
be added because the session size exceed the session limi
t ( 1048576 )
Mobyle.Execution.DRMAA : CRITICAL : DRMAA.py : L 281 : Thu, 22 Mar 2012 13:40:50 : error during
drmaa intitialization for job 4: code 2: unable to contact qmaster using port 6444 on host "musky"
Traceback (most recent call last):
File "mobyle/Src/Mobyle/Execution/DRMAA.py", line 277, in getStatus
s.initialize()
...
raise _ERRORS[code-1]("code %s: %s" % (code, error_buffer.value))
DrmCommunicationException: code 2: unable to contact qmaster using port 6444 on host "musky"](https://image.slidesharecdn.com/mobyleadministratorworkshop-121008100017-phpapp02/75/Mobyle-administrator-workshop-70-2048.jpg)