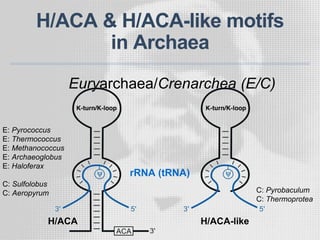
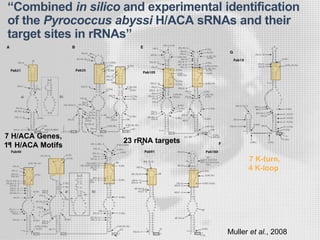
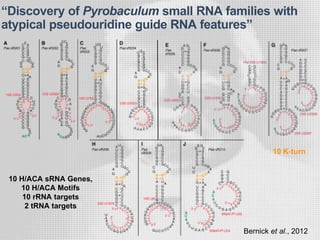
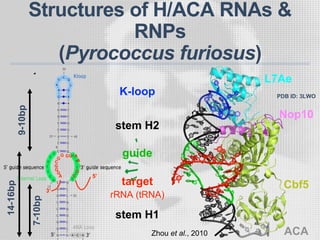
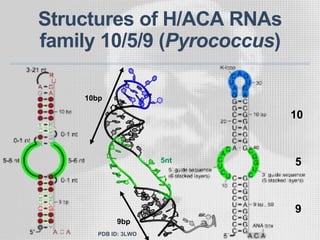
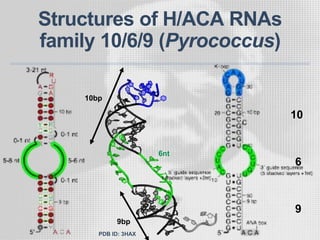
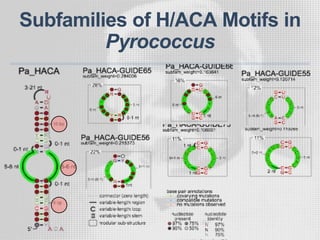
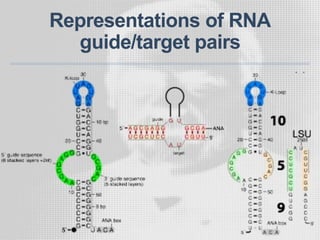
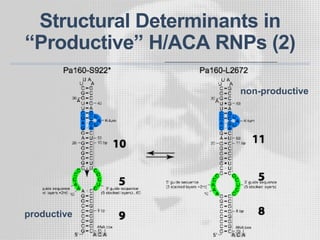
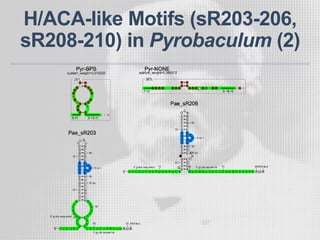
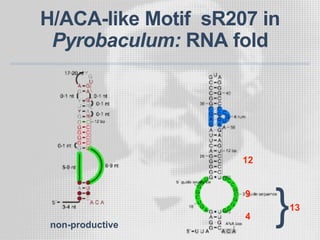
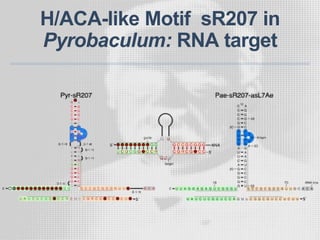
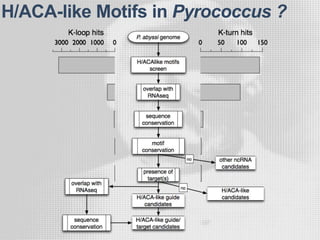
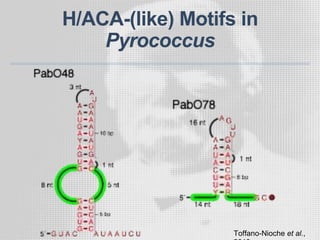
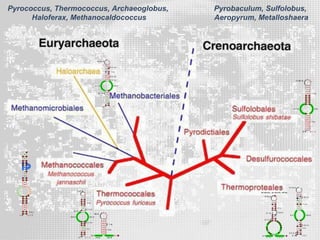
The document discusses the structure-function relationships of h/aca guide RNAs in archaea, focusing on their identification and roles in various archaeal species such as Pyrococcus and Pyrobaculum. It highlights the presence of both productive and non-productive h/aca and h/aca-like guide RNAs, and their variations across euryarchaea and crenarchaea. Key findings include the discovery of unique small RNA families and the potential alternative functions of these guide RNAs.