Unbiased peptide inhibitors of CD33 receptor
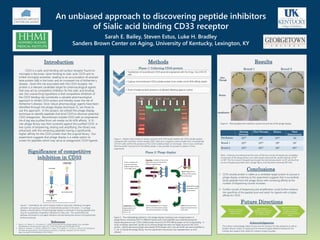
- 1. An unbiased approach to discovering peptide inhibitors of Sialic acid binding CD33 receptor Sarah E. Bailey, Steven Estus, Luke H. Bradley Sanders Brown Center on Aging, University of Kentucky, Lexington, KY Introduction CD33 is a sialic acid-binding cell surface receptor found on microglia in the brain. Upon binding to sialic acid, CD33 acts to inhibit microglial activation, leading to an accumulation of amyloid beta protein (Aβ) in the brain and an increased risk of Alzheimer’s disease. Given the risk associated with the CD33 receptor, the protein is a relevant candidate target for pharmacological agents that may act as competitive inhibitors for the sialic acid binding site. Our overarching hypothesis is that competitive inhibition of the CD33 binding site constitutes a valuable pharmacological approach to inhibit CD33 actions and thereby lower the risk of Alzheimer’s disease. Since robust pharmacologic agents have been identified through the phage display technique (1), we chose to use this approach. In this project, we utilized the phage display technique to identify peptides that bind CD33 to discover potential CD33 antagonists. Recombinant soluble CD33 with an engineered His-6 tag was purified from cell media via Ni-NTA affinity. A 12- mer phage library was then screened against the purified CD33. In two cycles of biopanning, eluting and amplifying, the library was enhanced, with the remaining peptides having a significantly higher affinity for the CD33 protein than the original library. Our experiment suggests that phage display is a viable option to screen for peptides which may serve as antagonistic CD33 ligands. Methods Results Significance of competitive inhibition in CD33 Figure 1. Uninhibited, the CD33 receptor binds to sialic acid, inhibiting microglial activation ad causing a build up of amyloid beta protein in the brain. In a phage display, a diverse library of bacteriophage display a multitude of varying peptides that may act as potential competitive inhibitors for sialic acid. This would block the pathway that leads to microglial inhibition and decreasing the amount of amyloid beta protein produced. Figure 2. Western blot testing of various amounts of Ni-NTA beads loaded with CD33 soluble protein compared with a positive control sample (U937 cells) and a negative control (media from untransfected CHO-K1 cells) confirm the presence of the CD33 soluble protein on the beads. Once it was confirmed that the protein had bound to the affinity beads, it was possible to proceed to phase 2 of the experiment. Transfection • Transfection of recombinant CD33 plasmid engineered with His-6 tag into CHO-K1 cells Protein Capture • Capture of recombinant CD33 soluble protein from media via NI-NTA affinity beads Testing • Proof of target protein presence via Western Blotting against control Phase 1: Collecting CD33 protein Round 1 Round 2 Figure 3. The methodology behind in vitro phage display screening over a target protein. A phage library containing 1x10^12 different twelve amino acid peptides was screened during the biopanning process over CD33 soluble protein bound to Ni-NTA affinity beads via His-6 tag binding. A series of washes were then carried out to remove phage that were not tightly bound to the CD33 protein. Specifically bound phage were eluted off the beads with a low pH buffer and were amplified in E. coli to rebuild the phage library. For this experiment, the process was repeated twice as time allowed. Figure 4. Titering plates from selection rounds one and two of the phage display. Mkr + ctrl 15 uL 10 uL -ctrl Mkr Phase 2: Phage display Starting Diversity Flow Through Elution Total Enhancement Pre-Screen 1013 107 1012 10 Round 1 1012 1010 106 107 Round 2 1010 1010 106 1011 Table 1. Diversity and enhancement for each of the two rounds of biopanning completed. A prescreen of the phage library over naïve beads enhanced the started diversity of 1013 to 1012. The first round of biopanning brought the total enhancement to 107 and the final round of biopanning ended with a phage library that had been enhanced 1011 fold. Conclusions Future Directions 1. CD33 soluble protein is viable as a candidate target protein to pursue in phage display screening as this experiment suggests that it successfully binds peptides from the phage library with increasing affinity as the number of biopanning rounds increases. 2. Further rounds of biopanning and amplification could further enhance the specificity of the peptide pool and select for ligands with a higher affinity for CD33 Further rounds of biopanning and amplification Sequencing of peptide combinations with highest affinity for CD33 protein Sequence analysis to interpret common binding patterns with pharmacological agents Acknowledgments The authors acknowledge Sabita Dumre and Jim Simpson for lab assistance as well as Sanders-Brown Center on Aging and the Howard Hughes Medical Institute for the funding and support that made this research project possible. References 1. Nixon, A. E., Sexton, D. J., & Ladner, R. C. (2014). Drugs derived from phage display: From candidate identification to clinical practice. mAbs, 6(1), 73–85. http://doi.org/10.4161/mabs.27240 2. Malik, M., Simpson, J. F., Parikh I, Wilfred, B. R., Fardo, D. W., Nelson, P. T. & Estus, S. (2013) CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 33 (33), 13320-5. http://doi.org/10.1523/JNEUROSCI.1224-13.2013 Flow Through Elution Amplification