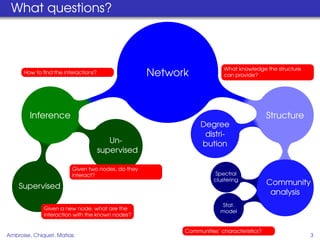
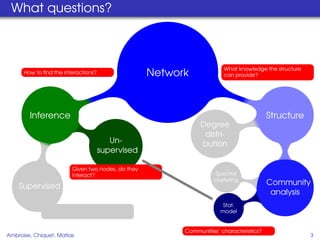
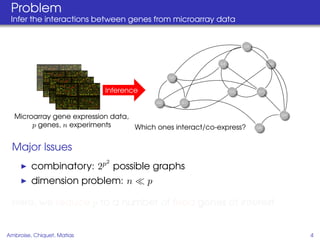
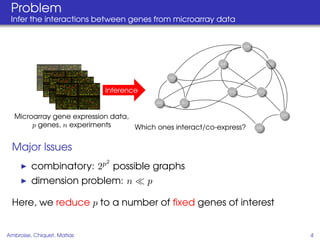
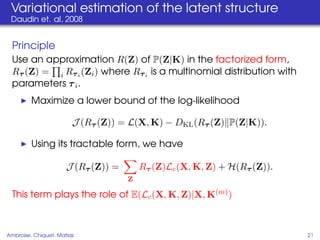
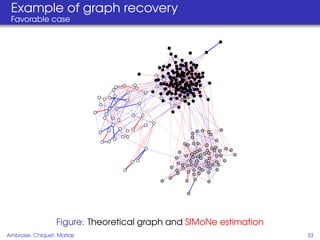
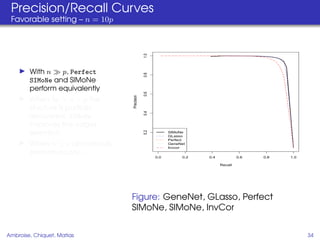
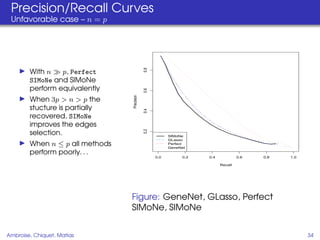
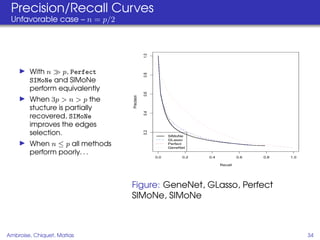
This document proposes a method for inferring sparse Gaussian graphical models with latent structure from biological data. It introduces a model that represents the concentration matrix as having a latent class structure, with vertices assigned to classes that determine their connectivity. An inference strategy alternates between estimating the latent class structure (E-step) and inferring the connectivity matrix (M-step). The method is evaluated on synthetic and breast cancer gene expression data.