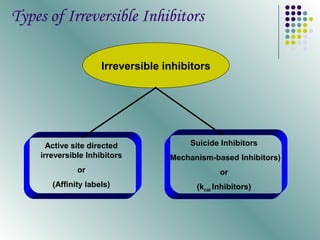
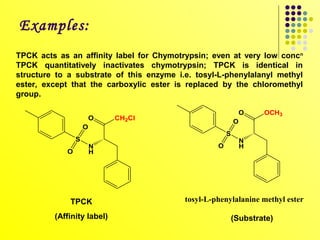
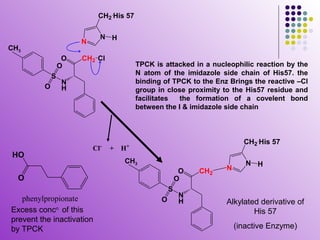
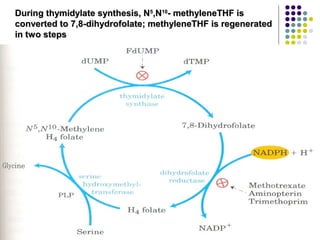
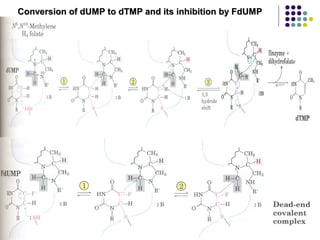
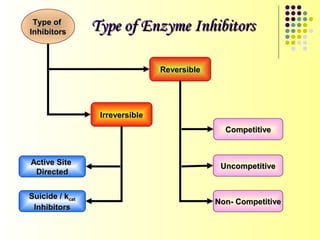
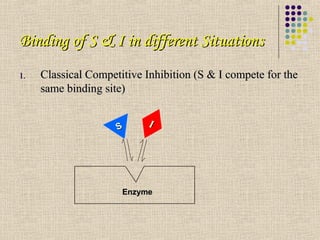
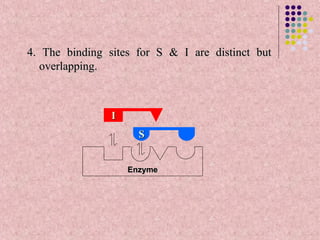
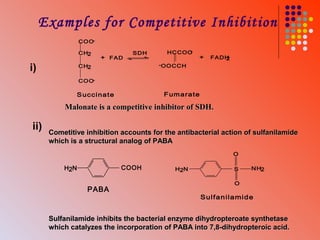
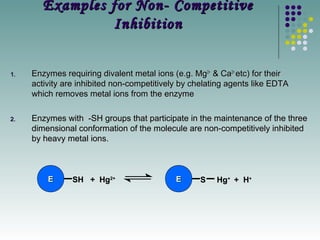
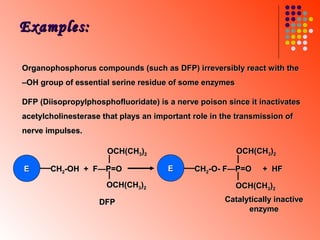
The document discusses different types of enzyme inhibition. It describes reversible and irreversible inhibition, and types of reversible inhibition including competitive, uncompetitive, and non-competitive inhibition. Competitive inhibitors bind to the active site of the enzyme and prevent substrate binding, while non-competitive inhibitors bind elsewhere and affect the enzyme's activity. The document provides examples and derivations of equations to describe these inhibition patterns and calculate inhibition constants.




![Competitive Inhibition
A competitive I combines with the free enzyme to form an EI
complex in a manner that prevents S binding
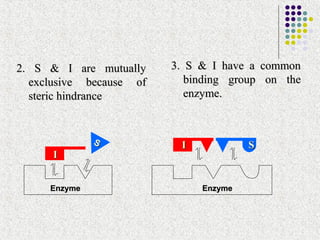
Binding of S & I is mutually exclusive
Inhibition can be reversed by increasing the concentration of S at a
constant [I]
Degree of inhibition will depend on the concentrations of S & I and
on the relative affinities of the enzyme for S & I](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-6-320.jpg)
![Derivation of velocity equation
k1 k2
E+S ES E+P
+ k-1
[E] [I]
I Ki = [EI]
Ki
[E] [I]
or [EI] = Ki
EI + S X No Reaction
In the steady state assumption
[E] [S] k-1 + k2
= = Km
[ES] k1
[E] [S]
[ES] = Km
v=k2[ES] ⇒ Vmax = k2 [E]T ⇒ Now [E]T = [E] + [ES] + [EI]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-12-320.jpg)
![Vmax = k2 ( [E] + [ES] + [EI] )
v k2 [ES] [ES]
= =
Vmax k2 ( [E] + [ES] + [EI] ) [E] + [ES] + [EI]
Putting the value of [ES] and [EI}
[E] [S]
v Km
Vmax =
[E] [E] [S] [E] [I]
+ +
Km Ki
[S]
v Km
=
Vmax [S] [I]
1 + Km + K
i](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-13-320.jpg)
![Multiplying by km both in the numerator and the
denominator
[S]
v
=
Vmax [I] Km
Km + [S] +
Ki
[S]
v
=
Vmax [I]
Km (1+ )+ [S]
+
Ki )](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-14-320.jpg)
![In the presence of a competitive inhibitor Km increases
Vmax unchanged
v [S]
=
Vmax Kmapp + [S]
[I]
Where Kmapp = Km (1+
Ki )
Vmax
v
No inhibitor
½ Vmax
+ C Inhibitor
Km Kmapp [s]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-15-320.jpg)
![[I]
Lineweaver Burk plot 1 Km ( 1+ Ki ) 1 1
=
v Vmax [S] + Vmax
[I] ) [I]2
+ i
(1 K
Km x [I]1
e = Vma
Sl op
1
Km
1
Kmapp](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-16-320.jpg)
![Calculation of Ki
From slope of the double reciprocal plot in the presence of a C.
Inhibitor which is egual to
Km (1+ [I] )
Slope = Ki
Vmax
From Kmapp which is given by
[I]
Kmapp = Km (1+
Ki )](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-17-320.jpg)
![ A graphical method is preferred to direct substitution of
numbers to allow errors in individual determination to be
averaged out
From the replot of slope vs. [I]
Km Km
Slope = + [I]
Vmax VmaxKi
Kmapp
Slope =
Vmax
Km
Slope =
Vmax Ki
Km
- Ki
Vmax
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-18-320.jpg)
![ From replot of Kmapp Vs. [I]
Km + Km [I]
Kmapp =
Ki
Kmapp
Km
Slope =
Ki
- Ki Km
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-19-320.jpg)
![ From Dixon’s plot Km [I] 1 (1+ Km )
1 = + [S]
v Vmax[S] Ki Vmax
IInc
nc
re
rea
as
[S]1
siin
ngg[
[SS]
]
1
Km Ki
v
x
[S] [S]
e = Vm
a
1 Km p
2
(1+ ) Slo
Vmax [S]
1 [S] = ∞
Vmax Slope = 0
[I]
[S]
- Ki (1+ Km )](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-20-320.jpg)

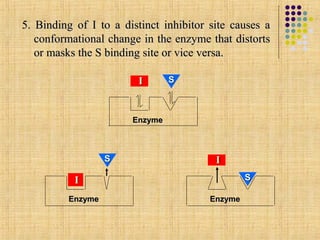
![ A NC I doesn’t affect the Km because the binding of I does not
block S binding or vice-versa
I effectively lowers the concentration of active enzyme and
hence decreases the apparent Vmax
since there is no competition between S & I, the inhibition is not
reversed by increasing the [S]
S
Enzyme Enzyme
S
I I
Enzyme Enzyme](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-22-320.jpg)
![k2
E+S ES E+P [E] [S] [EI] [S]
Ks Ks = [ES] =
+ + [ES]
I I
Replacing Ks with Km
Ki
Ki [ES] =
[E] [S]
Km
EI + S ESI `
Ks
Vmax
Vmax i
v No inhibitor
½ Vmax
+ NC Inhibitor
½ Vmax i
Vmax = Decreases.
Km = Unchanged
Km [s]→](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-24-320.jpg)
![ Lineweaver – Burk Plot
1 Km 1 + 1
=
v Vmaxi [S] Vmaxi
[I]2
m
K
i
ax
m
V
e=
[I]1
op
1/v
Sl
1 No Inhibitor
Both slope & Intercept =
Intercept Vmaxi
Increased By
the factor Km
Slope =
(1+[ I ] ) Vmax
1
Ki
Km 1
Intercept =
Vmax
1/[s]→](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-25-320.jpg)
![Calculation of Ki
i) From the slope of the reciprocal plot
ii) from the intercept of the reciprocal
plot Km Km [I]
Slope = +
iii) from replot of slope of the reciprocal Vmax Vmax Ki
plot vs [ I ]
Slope
In partial NC inhibition
this plot is hyperbolic
Km
- Ki
Vmax
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-26-320.jpg)
![iv. Replot of intercept of the primary plot in the presence
of a NC I vs [I] is linear
1 1 [I]
Intercept = +
Vmax Vmax Ki
Intercept
In partial NC inhibition
this plot is hyperbolic
1
- Ki
Vmax
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-27-320.jpg)
![v. Dixon’s Plot
A plot of 1/v vs [I] will be linear at
fixed [E] and [S] for NC inhibition
[S]1
[S]2
1/v Km 1 1
Slope = ( Vmax [S] + Vmax
) Ki
Km 1
- Ki
(
Intercept = Vmax [S] + Vmax )
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-28-320.jpg)
![Uncompetitive Inhibition
I doesn't bind to the free E rather it binds to the ES complex
the binding of an UC I is presumed to cause structural distortion
of the active site making the enzyme catalytically inactive
the binding of S could cause a conformational change in the E
thereby revealing an I binding site
Inhibition can’t be reversed by increasing the [S] since I doesn't
compete with S for the same binding site
S
Enzyme
Enzyme
S
I
Enzyme](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-29-320.jpg)
![ UC Inhibition is rare in single-substrate reactions.
for e.g. Inhibition of intestinal alkaline phosphatase by L-
phenylalanine. It is common in multisubstrate reactions
E+S ES E+P
+
I
[E] [S]
[ES] = Km
[E] [S] [I]
ESI [ESI] =
Km Ki](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-30-320.jpg)
![ The equilibria show that at any [I] an infinitely high [S] will not
drive all the enzyme to ES form; some non productive ESI complex
will always be present. Consequently an UC I will decrease the V max
An UC I will also decrease the Kmapp because the reaction
ES + I ESI removes some ES causing the reaction
E+S ES to proceed to the right](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-31-320.jpg)
![v [s]
=
Vmax Km [s]
[I] [I]
+
(1+ ) (1+ )
Ki Ki
The equation can also be written as
Vmax
v [s]
=
Vmaxi Kmapp +[s]
Vmax i
v ½ Vmax No inhibitor Vmax
+ UC Inhibitor Where Vmaxi = [I]
½ Vmax i (1+ )
Ki
Vmax = Decreases
Km = Decreases
Km
Kmapp=
[I]
Km [s]→ (1+ )
Kmapp Ki](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-32-320.jpg)
![Lineweaver Burk plot
[I]
1 Km 1 1 (1+ )
= + Ki
v Vmax [S] Vmax
Slope remains
Unchanged &
Intercept
Inc
Increases By
re
as
the factor
in
(1+[ I ] ) [I]2
g[
[I]1
I]
Ki 1/v
No I
1/Vmaxi
Incase of UC Inhibition Ki
Km is that concn of I which
Slope = halves the value of both
Vmax Vmax and Km
1/Vmax
1/[s]→
-1/Kmapp -1/Km](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-33-320.jpg)
![Calculation of Ki
i) From the slope of the reciprocal plot [I]
1 1 (1+ )
ii) From the Km app = Ki
Vmaxi Vmax
iii) From replot of 1/Vmaxi vs [ I ]
1 1 1 [I]
= +
Vmaxi Vmax VmaxKi
1/Vmaxi
1
Slope =
VmaxKi
1
- Ki
Vmax
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-34-320.jpg)
![iv. From replot of 1/Km appvs [I]
[I]
1 1 (1+ )
= Ki
Km app Km
1 1 1 [I]
= +
Kmapp Km KmKi
1/Kmapp
1
Slope =
KmKi
1
- Ki
Km
[I]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-35-320.jpg)
![The equation for Dixon’s plot is
iv. Dixon’s Plot
Km
1 Km [I] 1 (1+ )
= + [S]
v Vmax Ki Vmax
In c
1 i
xK
rea
= ma
sin
1/v o pe V
Sl
g[
1 Km
(1+ )
S]
Vmaxi [S]
∞
]=
[S
1/Vmax
[I]→
Km -Ki
-Ki (1+ )
[S]](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-36-320.jpg)

![To distinguish between irreversible & NC Inhibition
t or
ib i
In h
r
to
t or
bi
no
hi
i bi
In
o l)
In h
Vmax
C
nt r
N
+
ble
(Co
rsi
ve
Ir re
[E]i
[E]T→](https://image.slidesharecdn.com/enzymeinhibitions-130405020003-phpapp02/85/Enzyme-inhibitions-39-320.jpg)