This document summarizes a ddPCR supermix containing reagents for droplet digital PCR assays using EvaGreen dye. Key points:
1) The supermix is a ready-to-use reaction cocktail containing polymerase, nucleotides, buffer, and EvaGreen dye for partitioning DNA into droplets and performing PCR for quantification of DNA targets.
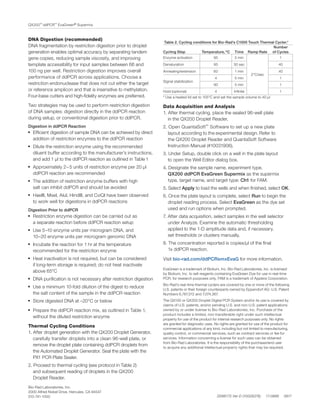
2) The supermix supports conventional cycling protocols and is compatible with UNG decontamination protocols.
3) The document provides instructions for setting up ddPCR reactions with the supermix, performing thermal cycling, and analyzing the droplet data.