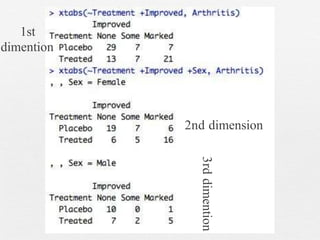
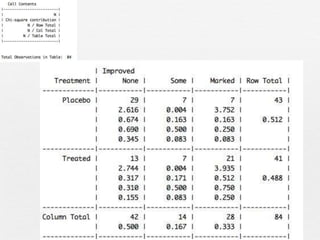
This document discusses various methods for summarizing categorical data in R, including table(), summary(), xtabs(), ftable(), prop.table(), and CrossTable(). It provides examples using the built-in Arthritis dataset to demonstrate how to index data frames, check factor levels and lengths, create one-variable and cross tables, calculate proportions, and more. Interactive exercises are included throughout for the reader to practice these methods on the Arthritis data.








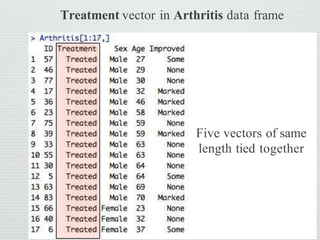
![Arthritis[1:17 , ]
Extract 1st to 17th rows Show all columns
Indexing: extraction of data from
data frame
Don’t forget comma
Colon in between](https://image.slidesharecdn.com/cdappt-240312130512-44a157d7/85/categorical-data-analysis-in-r-studioppt-pptx-10-320.jpg)