Cameron_Locker_variants_final_poster1
•Download as PPTX, PDF•
0 likes•363 views
The document describes a study that aimed to validate insertions and deletions (INDELs) identified in the Genome in a Bottle (GIAB) reference standard by comparing calls made by FreeBayes and GATK variant callers. Researchers analyzed sequencing data from the NA12878 genome using the two callers and GIAB, generating 7 variant lists. They selected 150 random INDELs from each list and designed primers to amplify the regions, then validated the variants using MiSeq sequencing. The results helped improve understanding of the variant callers and the GIAB standard for INDEL calls.
Report
Share
Report
Share
Recommended
Statistical methods for off-target variant genotyping on Affymetrix' Axiom Ar...

Teresa Webster, Bioinformatics, Affymetrix.Automated clustering and calling of ""Off-Target Variants,"" atypical cluster patterns caused by secondary variants in the probe hybridization region.
Best practices for genotyping analysis of plant and animal genomes with Affym...

Ali Pirani, Bioinformatics, Affymetrix.Overview of the best workflow to get from data to results in the shortest time.
Multigenic (mechanistic) biomarkers

SEQC2, Advancing Precision Medicine and Cancer Genomics,
Bethesda, MD, September 13‐14, 2016
Recommended
Statistical methods for off-target variant genotyping on Affymetrix' Axiom Ar...

Teresa Webster, Bioinformatics, Affymetrix.Automated clustering and calling of ""Off-Target Variants,"" atypical cluster patterns caused by secondary variants in the probe hybridization region.
Best practices for genotyping analysis of plant and animal genomes with Affym...

Ali Pirani, Bioinformatics, Affymetrix.Overview of the best workflow to get from data to results in the shortest time.
Multigenic (mechanistic) biomarkers

SEQC2, Advancing Precision Medicine and Cancer Genomics,
Bethesda, MD, September 13‐14, 2016
Monitoring the quality of data in the clinical use of pathogen genomes

Monitoring the quality of data in the clinical use of pathogen genomesHealth Informatics New Zealand
Dr Tom Conway, HiNZ 2015 Conference, ChristchurchThe Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...

The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...CrimsonpublishersCJMI
The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of Mycobacterium spp. from Smear Positive and Negative Sputum Samples by Gülnur Tarhan in ohesive Journal of Microbiology & Infectious DiseaseGene expression profile analysis of human hepatocellular carcinoma using sage...

Use of SAGE technique In Research
Development of a high throughput workflow for genotyping CFTR mutations

Cystic Fibrosis is an autosomal recessive genetic disease that is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which has important roles in ion exchange.
Bioinformatics in dermato-oncology

Bioinformatics methods in dermato-Oncology
VII Symposium Internacional Avances Oncologic Dermatology
Valencia (Spain) 24th October, 2014
The server of the Spanish Population Variability

DNA Day
Hospital Universitario La Paz, Madrid, Spain April 28th, 2014
The first server of the Spanish Population Variability.
Freely available: http://ciberer.es/bier/exome-server/
See alse related tools:
BiERapp: http://bierapp.babelomics.org (to help in the prioritization of disease genes)
TEAM: http://team.babelomics.org (to manage panels of genes for targeter resequencing based diagnostic)
Identification of antibiotic resistance genes in Klebsiella pneumoniae isolat...

Antibiotic resistant strains of pathogenic bacteria are a growing worldwide health problem. To effectively combat the spread of difficult-to-treat bacterial infections, rapid surveillance methods for detection of antibiotic resistance genes is required to monitor both bacterial isolates and metagenomic samples. Additionally, identification of potential new sources for different antibiotic resistance genes is critical. Both of these goals require tools that can be used for profiling of antibiotic resistance genes from various types of samples. Real-time PCR has proven to be effective for the detection of antibiotic resistance genes. Using PCR array technology, simultaneous detection of 87 prevalent and important antibiotic resistance genes is possible and should prove to be an effective method for antibiotic resistance monitoring. This allows for a more comprehensive profiling of antibiotic resistance genes than is possible using individual PCR assays.
Multiplex TaqMan Assays for Rare Mutation Analysis Using Digital PCR

Detection of rare mutations in tumor tissue and cell free DNA (cfDNA) allows for monitoring of tumor progression and regression for research purposes. cfDNA isolated from plasma combined with a sensitive detection method like digital PCR is non- invasive and enables earlier detection compared to conventional imaging techniques. Building on the TaqMan based Rare Mutation assay set for detection of rare mutations using digital PCR on the QuantStudio 3D Digital PCR System, we are now developing multiplex assays for simultaneous detection of several mutations. We selected relevant mutations in the EGFR and KRAS genes for our initial multiplex application: EGFR G719, EGFR exon 19 deletions, and
KRAS G12/G13. These mutations may have implications for potential future targeted therapy. Primers and probes of singleplex Rare Mutation Assays were reformulated to generate multiplex assays detecting the EGFR and KRAS mutations. All multiplex assays were tested on template composed of wild-type genomic DNA background mixed with mutant plasmid reflecting each of the mutations detected by the multiplex
assays. Initial experimental results were successful and showed excellent signal intensity and clear cluster separation when analyzed with the QuantStudio 3D AnalysisSuiteTM Cloud Software. The EGFR G719 mutations (COSM6239, COSM6253, COSM6252) were detected using a 3plex assay, EGFR exon 19 deletions (COSM12383, COSM12422, COSM12678, COSM6223, COSM6254, COSM6255) were detected using a 6plex assay, and KRAS G12/G13 mutations (COSM516,
COSM517, COSM518, COSM520, COSM521, COSM522, COSM527, COSM532) were detected using an 8plex. Multiplexing assays for three relevant mutation loci proved feasible and presents an efficient way to assess the presence and the percentage of mutations at these loci.
Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...

Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...Thermo Fisher Scientific
TaqMan genotyping assays are widely used to genotype CYP2D6, which encodes a major drug metabolizing enzyme.One man's *1 is another man's *13? Trouble with nomenclatures in personalized...

Slides for a talk at SUMMER SCHOOL: GENOMIC MEDICINE – Bridging research and the clinic, May 6 2016, Portoroz, Slovenia
Digging into thousands of variants to find disease genes in Mendelian and com...

IV DCEX symposium, PRBB, Barcelona, November 17th, 2015
More Related Content
What's hot
Monitoring the quality of data in the clinical use of pathogen genomes

Monitoring the quality of data in the clinical use of pathogen genomesHealth Informatics New Zealand
Dr Tom Conway, HiNZ 2015 Conference, ChristchurchThe Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...

The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...CrimsonpublishersCJMI
The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of Mycobacterium spp. from Smear Positive and Negative Sputum Samples by Gülnur Tarhan in ohesive Journal of Microbiology & Infectious DiseaseGene expression profile analysis of human hepatocellular carcinoma using sage...

Use of SAGE technique In Research
Development of a high throughput workflow for genotyping CFTR mutations

Cystic Fibrosis is an autosomal recessive genetic disease that is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which has important roles in ion exchange.
Bioinformatics in dermato-oncology

Bioinformatics methods in dermato-Oncology
VII Symposium Internacional Avances Oncologic Dermatology
Valencia (Spain) 24th October, 2014
The server of the Spanish Population Variability

DNA Day
Hospital Universitario La Paz, Madrid, Spain April 28th, 2014
The first server of the Spanish Population Variability.
Freely available: http://ciberer.es/bier/exome-server/
See alse related tools:
BiERapp: http://bierapp.babelomics.org (to help in the prioritization of disease genes)
TEAM: http://team.babelomics.org (to manage panels of genes for targeter resequencing based diagnostic)
Identification of antibiotic resistance genes in Klebsiella pneumoniae isolat...

Antibiotic resistant strains of pathogenic bacteria are a growing worldwide health problem. To effectively combat the spread of difficult-to-treat bacterial infections, rapid surveillance methods for detection of antibiotic resistance genes is required to monitor both bacterial isolates and metagenomic samples. Additionally, identification of potential new sources for different antibiotic resistance genes is critical. Both of these goals require tools that can be used for profiling of antibiotic resistance genes from various types of samples. Real-time PCR has proven to be effective for the detection of antibiotic resistance genes. Using PCR array technology, simultaneous detection of 87 prevalent and important antibiotic resistance genes is possible and should prove to be an effective method for antibiotic resistance monitoring. This allows for a more comprehensive profiling of antibiotic resistance genes than is possible using individual PCR assays.
Multiplex TaqMan Assays for Rare Mutation Analysis Using Digital PCR

Detection of rare mutations in tumor tissue and cell free DNA (cfDNA) allows for monitoring of tumor progression and regression for research purposes. cfDNA isolated from plasma combined with a sensitive detection method like digital PCR is non- invasive and enables earlier detection compared to conventional imaging techniques. Building on the TaqMan based Rare Mutation assay set for detection of rare mutations using digital PCR on the QuantStudio 3D Digital PCR System, we are now developing multiplex assays for simultaneous detection of several mutations. We selected relevant mutations in the EGFR and KRAS genes for our initial multiplex application: EGFR G719, EGFR exon 19 deletions, and
KRAS G12/G13. These mutations may have implications for potential future targeted therapy. Primers and probes of singleplex Rare Mutation Assays were reformulated to generate multiplex assays detecting the EGFR and KRAS mutations. All multiplex assays were tested on template composed of wild-type genomic DNA background mixed with mutant plasmid reflecting each of the mutations detected by the multiplex
assays. Initial experimental results were successful and showed excellent signal intensity and clear cluster separation when analyzed with the QuantStudio 3D AnalysisSuiteTM Cloud Software. The EGFR G719 mutations (COSM6239, COSM6253, COSM6252) were detected using a 3plex assay, EGFR exon 19 deletions (COSM12383, COSM12422, COSM12678, COSM6223, COSM6254, COSM6255) were detected using a 6plex assay, and KRAS G12/G13 mutations (COSM516,
COSM517, COSM518, COSM520, COSM521, COSM522, COSM527, COSM532) were detected using an 8plex. Multiplexing assays for three relevant mutation loci proved feasible and presents an efficient way to assess the presence and the percentage of mutations at these loci.
Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...

Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...Thermo Fisher Scientific
TaqMan genotyping assays are widely used to genotype CYP2D6, which encodes a major drug metabolizing enzyme.One man's *1 is another man's *13? Trouble with nomenclatures in personalized...

Slides for a talk at SUMMER SCHOOL: GENOMIC MEDICINE – Bridging research and the clinic, May 6 2016, Portoroz, Slovenia
Digging into thousands of variants to find disease genes in Mendelian and com...

IV DCEX symposium, PRBB, Barcelona, November 17th, 2015
What's hot (20)
Bioinformatics-driven discovery of EGFR mutant Lung Cancer

Bioinformatics-driven discovery of EGFR mutant Lung Cancer
EHA poster Genomic Analysis by MyAML with Chemotherapy

EHA poster Genomic Analysis by MyAML with Chemotherapy
Monitoring the quality of data in the clinical use of pathogen genomes

Monitoring the quality of data in the clinical use of pathogen genomes
The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...

The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of M...
Gene expression profile analysis of human hepatocellular carcinoma using sage...

Gene expression profile analysis of human hepatocellular carcinoma using sage...
Development of a high throughput workflow for genotyping CFTR mutations

Development of a high throughput workflow for genotyping CFTR mutations
Identification of antibiotic resistance genes in Klebsiella pneumoniae isolat...

Identification of antibiotic resistance genes in Klebsiella pneumoniae isolat...
Ruta graveolens extract induces dna damage pathways and blocks akt activation...

Ruta graveolens extract induces dna damage pathways and blocks akt activation...
Multiplex TaqMan Assays for Rare Mutation Analysis Using Digital PCR

Multiplex TaqMan Assays for Rare Mutation Analysis Using Digital PCR
Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...

Resolving false positive CYP2D6 genotype results: CYP2D7 variation is the cul...
One man's *1 is another man's *13? Trouble with nomenclatures in personalized...

One man's *1 is another man's *13? Trouble with nomenclatures in personalized...
Digging into thousands of variants to find disease genes in Mendelian and com...

Digging into thousands of variants to find disease genes in Mendelian and com...
Viewers also liked
GIAB Sep2016 Lightning Effective Depth Metric yves konigshofer_sera_care

GIAB Sep2016 Lightning Effective Depth Metric yves konigshofer_sera_care
Gente Tóxica...

Autor: Bernardo Stamateas
Aprender a relacionarse sanamente, identificando a las personas tóxicas, al igual que nuestro comportamiento tóxico ante o con los demás que nos rodean.
(La descarga puede ser muy lenta dado que tiene un peso de 56 megabytes)
agbt 2016 workshop lindsay

Tina Graves-Lindsay presentation at MGI-hosted workshop on reference genomes at AGBT 2016
«Diseño para todos» en la investigacion social sobre personas con discapacidad

«Diseño para todos» en la investigacion social sobre personas con discapacidadPedro Roberto Casanova
Autores: Mario Toboso-Martín: Instituto de Filosofía-CSIC / Jesús Rogero-García: Universidad Autónoma de Madrid
Los estudios sociales sobre la discapacidad han aumentado en número e importancia en España y otros países durante los últimos años. Sin embargo, la mayoría de fuentes de información y estudios disponibles no recogen de manera adecuada la realidad de un colectivo muy heterogéneo, que supone en la actualidad aproximadamente el 9 por ciento de la población española. La implementación de medidas sociales requiere de fuentes y estudios representativos que aporten información precisa acerca de estas personas. El objetivo de esta nota es identificar las principales difi cultades que se plantean a la hora de diseñar y llevar a la práctica metodologías de investigación social adecuadas hacia las personas con discapacidad, así como ofrecer propuestas y recomendaciones para avanzar hacia una investigación social más inclusiva, mediante los conceptos de accesibilidad y diseño para todos.La educación domiciliaria y hospitalaria en el nivel secundario 2016

Ministerio de Educación y Deportes de la Nación
La educación domiciliaria y hospitalaria en el nivel secundario. - 1a ed . - Ciudad Autónoma de Buenos Aires: Ministerio de Educación y Deportes, 2016.
“La obligatoriedad de la escuela secundaria representa la promesa y apuesta histórica de la sociedad argentina, como en otros momentos lo fue la escuela primaria, para la inclusión efectiva en la sociedad y la cultura de todos los adolescentes, jóvenes y adultos”
Viewers also liked (15)
GIAB Sep2016 Lightning Effective Depth Metric yves konigshofer_sera_care

GIAB Sep2016 Lightning Effective Depth Metric yves konigshofer_sera_care
«Diseño para todos» en la investigacion social sobre personas con discapacidad

«Diseño para todos» en la investigacion social sobre personas con discapacidad
La educación domiciliaria y hospitalaria en el nivel secundario 2016

La educación domiciliaria y hospitalaria en el nivel secundario 2016
Similar to Cameron_Locker_variants_final_poster1
Enabling CNV Studies from Single Cells Using Whole Genome Amplification and L...

DNA copy number variations (CNVs) play an important role in the pathogenesis and progression of cancer. While array comparative genomic hybridization (aCGH) has generally been used to identify CNVs in the whole genome, next-generation sequencing (NGS) provides an opportunity to characterize CNVs genome-wide with unprecedented resolution, even at the single cell level.
However, CNV detection in single cells is faced with various challenges, such as incomplete genome coverage, introduction of sequence errors, GC bias and false positives.
In this new poster, we show a method for capturing the entire genomic complexity of a single cell, overcoming these challenges and ensuring accurate detection of CNVs.
Rapid and accurate Cancer somatic mutation profiling with the qBiomarker Soma...

QIAGEN has developed real-time PCR-based qBiomarker Somatic Mutation PCR Arrays for pathway- and disease-focused mutation profiling. By combining allele-specific amplification and 5' hydrolysis probe detection, the PCR assays on these arrays detect as little as 0.01% somatic mutation in a background of wild-type genomic DNA. These assays have consistent and reliable mutation detection performance in different sample types (including fresh, frozen, or formalin-fixed samples), and with varying sample quality. In application examples, the PCR-based mutation detection results are consistent with Pyrosequencing results for the same samples. The qBiomarker Somatic Mutation PCR Arrays, combining laboratory-verified assays, comprehensive content, and integrated data analysis software, are highly suited for identifying somatic mutations in the context of biological pathways and diseases.
A novel method for building custom ampli seq panels using optimized pcr primers 

A novel method for building custom ampli seq panels using optimized pcr primers Thermo Fisher Scientific
AmpliSeq™ is a next generation sequencing library preparation method for targeted re-sequencing that utilizes highly multiplexed PCR to amplify regions of interest. A key to successful AmpliSeq libraries is the primer panel used for target amplification. Until now primers have been available as pre-assembled ready-to-use panels, or as custom made-to-order panels. We describe a new process for creating customized panels consisting of optimized and verified PCR primers. The primer sets are available as whole genes (i.e., all of the primers needed to create libraries that cover the entire coding regions of genes) and are selectable on the ampliseq.com website by either uploading gene lists or choosing genes from disease research areas.
We show NGS sequencing data from 10 disease research-oriented panels, including newborn screening research and inherited cancer research, assembled from individual pre-verified gene sets. Panel performance data include coverage uniformity, reproducibility, and sensitivity and positive predictive value of variant calling. To demonstrate flexibility of panel content and performance, the coverage uniformity of the 59 genes recommended by the American College of Medical Genetics and Genomics for reporting of incidental findings (ACMG59) was evaluated by themselves and with up to 135 additional genes and shown to be ≥ 97% in all contexts. We also demonstrate the robustness of this method using a variety of sample types (fresh, frozen, and dried blood, cheek swabs) with both manual and fully automated library preparation methods. For Research use only. Not for use in diagnostic procedures.
SAGE- Serial Analysis of Gene Expression

It will give Idea About SAGE- Serial Analysis of Gene Expression
Visual Exploration of Clinical and Genomic Data for Patient Stratification

Talk presented at the Simons Foundation Biotech Symposium "Complex Data Visualization: Approach and Application" (12 September 2014)
http://www.simonsfoundation.org/event/complex-data-visualization-approach-and-application/
In this talk I describe how we integrated a sophisticated computational framework directly into the StratomeX visualization technique to enable rapid exploration of tens of thousands of stratifications in cancer genomics data, creating a unique and powerful tool for the identification and characterization of tumor subtypes. The tool can handle a wide range of genomic and clinical data types for cohorts with hundreds of patients. StratomeX also provides direct access to comprehensive data sets generated by The Cancer Genome Atlas Firehose analysis pipeline.
http://stratomex.caleydo.org
Mastering RNA-Seq (NGS Data Analysis) - A Critical Approach To Transcriptomic...

This workshop will address critical issues related to Transcriptomics data:
Processing raw Next Generation Sequencing (NGS) data:
1. Next Generation Sequencing data preprocessing:
Trimming technical sequences
Removing PCR duplicates
2. RNA-seq based quantification of expression levels:
Conventional pipelines (looking at known transcripts)
Identification of novel isoforms
Analysis of Expression Data Using Machine Learning:
3. Unsupervised analysis of expression data:
Principal Component Analysis
Clustering
4. Supervised analysis:
Differential expression analysis
Classification, gene signature construction
5. Gene set enrichment analysis
The workshop will include hands-on exercises utilizing public domain datasets:
breast cancer cell lines transcriptomic profiles (https://genomebiology.biomedcentral.com/articles/10.1186/gb-2013-14-10-r110),
patient-derived xenograft (PDX) mouse model of tumor and stroma transcriptomic profiles (http://www.oncotarget.com/index.php?journal=oncotarget&page=article&op=view&path[]=8014&path[]=23533), and
processed data from The Cancer Genome Atlas samples (https://cancergenome.nih.gov/).
Team: The workshops are designed by the researchers at the Tauber Bioinformatics Research Center at University of Haifa, Israel in collaboration with academic centers across the US. Technical support for the workshops is provided by the Pine Biotech team. https://edu.t-bio.info/a-critical-approach-to-transcriptomic-data-analysis/
Targeted Single Cell Sequencing for Accurate Mutation Detection in Heterogene...

Targeted Single Cell Sequencing for Accurate Mutation Detection in Heterogeneous Cellular Populations
“MS-Extractor: An Innovative Approach to Extract Microsatellites on „Y‟ Chrom...

Simple Sequence Repeats (SSR), also known as Microsatellites, have been extensively used as
molecular markers due to their abundance and high degree of polymorphism. The nucleotide sequences of
polymorphic forms of the same gene should be 99.9% identical. So, Microsatellites extraction from the Gene is
crucial. However, Microsatellites repeat count is compared, if they differ largely, he has some disorder. The Y
chromosome likely contains 50 to 60 genes that provide instructions for making proteins. Because only males
have the Y chromosome, the genes on this chromosome tend to be involved in male sex determination and
development. Several Microsatellite Extractors exist and they fail to extract microsatellites on large data sets of
giga bytes and tera bytes in size. The proposed tool “MS-Extractor: An Innovative Approach to extract
Microsatellites on „Y‟ Chromosome” can extract both Perfect as well as Imperfect Microsatellites from large
data sets of human genome „Y‟. The proposed system uses string matching with sliding window approach to
locate Microsatellites and extracts them.
ASHG 2015 - Redundant Annotations in Tertiary Analysis

After obtaining genetic variants from next generation sequencing data, a precursory step in tertiary analysis is to annotate each variant with available relevant information. There is no standardized compendium for this purpose; researchers instead are required to compile data from a motley of annotation tools and public datasets. These sources for annotation are independently maintained, and accordingly there is limited concordance between their reported contents. The choice of annotation datasets thus has a direct and significant impact on the results of the analysis.
Custom AmpliSeq™ Panels for Inherited Disease Research from Optimized, Invent...

Custom AmpliSeq™ Panels for Inherited Disease Research from Optimized, Invent...Thermo Fisher Scientific
Next generation sequencing (NGS) refers to the parallel sequencing of hundreds to
millions of DNA fragments. The consequent increase in throughput and decrease in
price has led to an explosion of genetic information for a variety of organisms. Ion
AmpliSeq™ library preparation is a multiplexed PCR-based method for amplifying
and preparing specific regions of interest in a genome for sequencing on Ion
Torrent™ NGS instruments. This library preparation method can accommodate
from dozens to many thousands of primer pairs in single tube reactions, and can
utilize from one to many primer pools for maximum flexibility. Primers are available
as pre-assembled ready-to-use panels or as custom made-to-order panels.Integration of single molecule, genome mapping data in a web-based genome bro...

Sequence, Finishing and the Future Conference (SFAF 2015) Poster submission. Santa Fe, New Mexico.
Poster describes the gEVAL browser, and the integration of genome/optical map data for use in evaluating/curation of genome assemblies. Human, Mouse, Zebrafish, Pig, Helminth, Chicken.
Whole Transcriptome Analysis of Testicular Germ Cell Tumors

Next generation sequencing of the whole transcriptome enables high resolution measurement of gene expression activity in different tissue and cell types. This methodology provides an in depth study of known transcripts and depending on the data analysis, allows identification of additional transcript types such as transcript variants, fusion transcripts, and small and long ncRNAs.
In this study we performed RNA-Seq using the Ion Torrent™ sequencing platform to compare the expression profile of testicular germ cell cancers (seminoma type, n=3) and normal testis (n=3). Using Partek Flow® 3.0 and TopHat/BowTie or Star aligners, we aligned the reads to the human genome and mapped sequences to the RefSeq database. Differentially expressed genes were identified and screened with additional germ cell tumors.
PCA analysis showed clear separation of the two sample types indicating biological differences. List of differentially expressed genes generated from TopHat/Bowtie and Star were similar. We identified a large number of genes that were up and down regulated with high degree of significance (p<0.01,>2X FC (fold change)). These included genes related to testicular tissue type, stem cell pluripotency (NANOG; POU5F1) and proliferation (KRAS, CCND2).
In addition, a number of differentially expressed noncoding RNAs were identified (SNORD12B, XIST). The method was validated on a small set of genes (n=20) using qPCR (TaqMan® Assays) and were found to be correlated. We used the OpenArray® platform to quickly and quantitatively screen 102 differentially expressed genes and 10 endogenous control genes across a number of different testicular germ cell cancer types.
We used a complete work flow solution from sample prep to NGS to qPCR to compare the expression profile of normal testis and seminoma type germ cell tumors. From the NGS experiments we identified a large number of differentially expressed genes for qPCR screening with samples from different types of germ cell tumors. Results from these screening studies will be presented.
Similar to Cameron_Locker_variants_final_poster1 (20)
Enabling CNV Studies from Single Cells Using Whole Genome Amplification and L...

Enabling CNV Studies from Single Cells Using Whole Genome Amplification and L...
Rapid and accurate Cancer somatic mutation profiling with the qBiomarker Soma...

Rapid and accurate Cancer somatic mutation profiling with the qBiomarker Soma...
A novel method for building custom ampli seq panels using optimized pcr primers 

A novel method for building custom ampli seq panels using optimized pcr primers
A High Throughput TaqMan CFTR Mutation Genotyping Workflow

A High Throughput TaqMan CFTR Mutation Genotyping Workflow
Visual Exploration of Clinical and Genomic Data for Patient Stratification

Visual Exploration of Clinical and Genomic Data for Patient Stratification
Mastering RNA-Seq (NGS Data Analysis) - A Critical Approach To Transcriptomic...

Mastering RNA-Seq (NGS Data Analysis) - A Critical Approach To Transcriptomic...
Targeted Single Cell Sequencing for Accurate Mutation Detection in Heterogene...

Targeted Single Cell Sequencing for Accurate Mutation Detection in Heterogene...
“MS-Extractor: An Innovative Approach to Extract Microsatellites on „Y‟ Chrom...

“MS-Extractor: An Innovative Approach to Extract Microsatellites on „Y‟ Chrom...
ASHG 2015 - Redundant Annotations in Tertiary Analysis

ASHG 2015 - Redundant Annotations in Tertiary Analysis
Custom AmpliSeq™ Panels for Inherited Disease Research from Optimized, Invent...

Custom AmpliSeq™ Panels for Inherited Disease Research from Optimized, Invent...
Integration of single molecule, genome mapping data in a web-based genome bro...

Integration of single molecule, genome mapping data in a web-based genome bro...
Whole Transcriptome Analysis of Testicular Germ Cell Tumors

Whole Transcriptome Analysis of Testicular Germ Cell Tumors
Cameron_Locker_variants_final_poster1
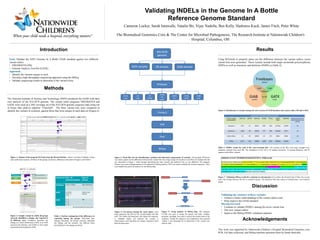
- 1. Validating INDELs in the Genome In A Bottle Reference Genome Standard Cameron Locker, Sarah Imawalle, Natalie Bir, Vijay Nadella, Ben Kelly, Harkness Kuck, James Fitch, Peter White The Biomedical Genomics Core & The Center for Microbial Pathogenesis, The Research Institute at Nationwide Children's Hospital, Columbus, OH Methods The National Institute of Science and Technology (NIST) produced the GIAB with their own analysis of the NA12878 genome. The variant caller programs FREEBAYES and GATK were used on a 30X coverage set of the NA12878 genome sequence data using our in house data analysis pipeline “Churchill". The three variant lists were compared to identify the variants in common, against those that were unique to each data set (Figure 6) ResultsIntroduction Goal: Validate the NIST Genome In A Bottle GIAB standard against two different variant callers • FREEBAYES (FB) • Genome Analysis Tool Kit (GATK) Approach: • Identify the variants unique to each • Develop a high throughput sequencing approach using the MiSeq • Validate sequencing results to determine if the variant exists. Figure 2: Sample variant in which all groups correctly identified a change, but reported it in different ways. Freebayes describes the change as a block substitution, GATK as an SNP, insertion and deletion, and GIAB as four single nucleotide polymorphisms (SNP). Figure 1: Output of the program IGVtools from the Broad Institute. Allows viewing of multiple variant calls at the same location. All three of the groups are shown, differences described in Figure 2 and Table 1. Table 1: Further explanation of the differences in reporting among the groups. Individual base changes described. All groups correctly identified the change and the location, but differed when describing how the change occurred. Figure 3: Work flow for the identification, isolation and laboratory preparation of variants. The program RTGtools was used to parse out the differences between the variant lists. By using a range for location it can filter out variants like the one described in Figure 1 where despite describing the same variant, the method to do so was different. The program Primer3 was used to design primers to be ordered for MiSeq analysis. PCR was used to amplify the desired sequences. They were loaded into pools for analysis by the MiSeq data. Figure 5: Fastq analysis of MiSeq data. The program FLASh was used to merge the paired end reads. Another program, cutAdapt, was used to remove the primer ends of the sequence. Finally the sequence was compared to the expected variant it was designed for to determine if the variant was indeed valid. Using RTGtools to properly parse out the difference between the variant callers, seven variant lists were generated . These variants include both single nucleotide polymorphisms (SNPs) as well as insertions and deletions (INDELs) (Table 2). Figure 6: Distribution of variants among the cross-section of GIAB database and variant callers FB and GATK. Table 2: INDEL counts for each of the cross-sectional lists. 150 variants (if the files were large enough) were randomly selected from each file. The breakdown of the 150 is 70 random insertions, 70 random deletions, and 10 random multi allelic variants. Figure 4: Gel process during the ‘pool’ phase. Each band represents the DNA of the variant loaded into that well. The further the band goes, the lighter the sequence is. Multiple bands can indicate the variant is heterozygous such that there are unique sequences from both chromosomes. Validation of chr2: 79133830 CACACACACCCTAT>C GIAB variant GGTGGCACAGATAAGGACACAGTAGTCATGAGCTTTTGCCCACAGTAAACTGGATGATTACTGAAAGAAAGGGAGGCTGACAAGGAGAG CCTGTGATTAAGGTAGAAAAGGTTTTCAACCAGGGCCCTTTCAAGCAGCACTGAGAACATTTCAGCTTCTTCCTTCCAGCCTTGGAGAG GAAAGTACACACACACACACACACACACACACACACACACACACACACACACACACCCTATCTTTTTTTTTCTTTTGACTAAAGACAGA TGATGACATGGTTGACCAGTATTCACACACACTCAAAGAAGTTAAATGCTTTTTAGCTGACAGTCATCTCAAATCCTTCTAGAAAACAA CACAAAATACTTTATGTGATTTGCTGGTCACTTCACTGTTTAGCCC Figure 7: Validating MiSeq results for a deletion on chromosome 2. In yellow, the forward read, in blue, the reverse read. The overlap between the two is colored in green. The deletions from this region of chromosome 2 are found in purple. GIAB Freebayes GATK Block Substitution Insertion and Deletion SNPs Variant List Ref > Alt GTACCA>GCGGCG Type of Variant Freebayes TACCA > CGGCG Block Substitution GATK G> GCGG GTAC > G A > G Insertion and Deletion GIAB T > C A > G C > G A > G SNP All correctly report the same result Acknowledgements This work was supported by Nationwide Children’s Hospital Biomedical Genomics core. PCR, Gel data collection, and MiSeq machine operation done by Sarah Imawalle. Discussion Validating the existence of these variants: • Achieve a better understanding of the variant callers used • Help improve the GIAB standard Moving Forward: • Continue to validate INDELs among the seven variant lists • Test new variant callers • Improve the MiSeq INDEL validation pipeline