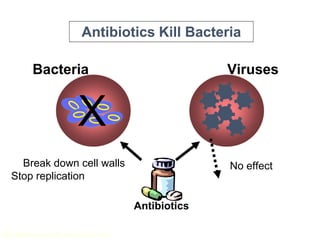
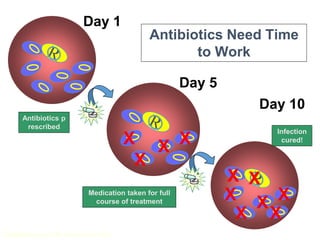
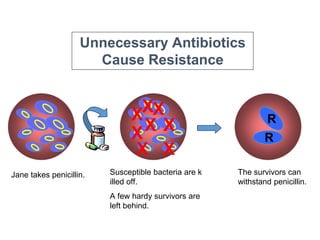
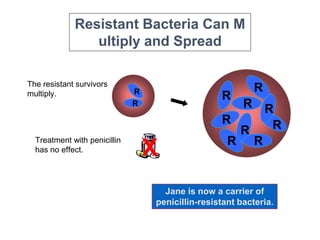
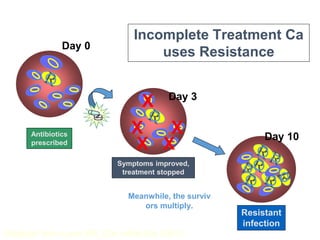
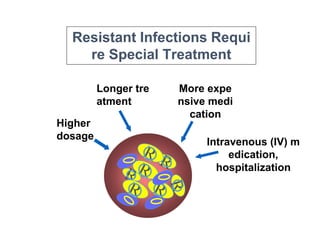
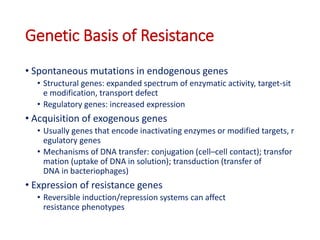
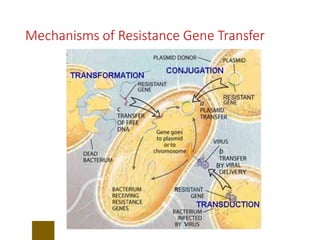
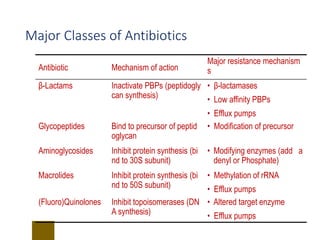
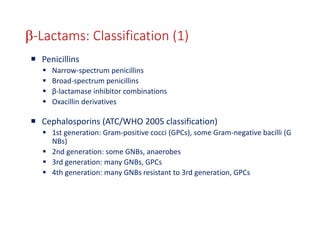
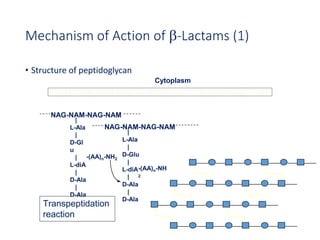
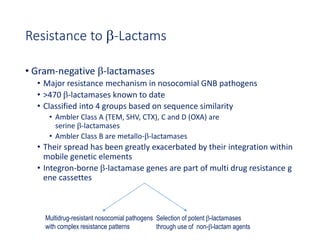
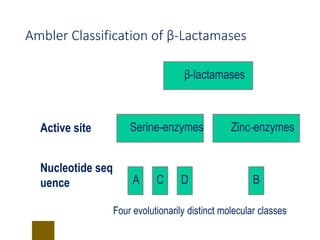
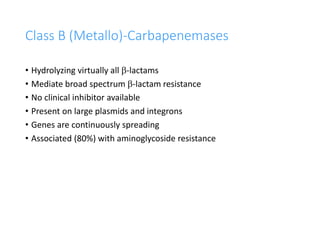
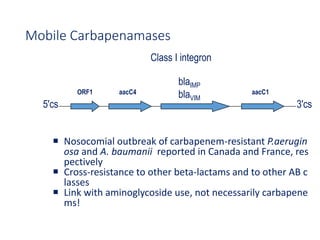
This document discusses antibiotic resistance and how it develops. It notes that bacteria can become resistant to antibiotics through spontaneous mutations, acquiring resistance genes from other bacteria, or incomplete treatment that allows resistant strains to survive and multiply. When antibiotics are overused, it creates selective pressure that promotes the spread of resistance. This threatens our ability to treat bacterial infections effectively as more infections become difficult or impossible to treat.