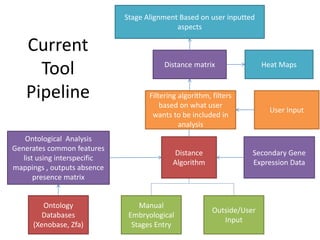
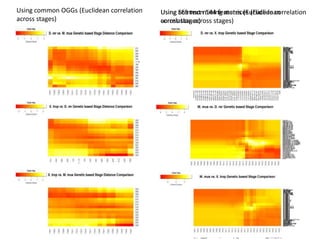
This document describes an embryo stage alignment tool that compares gene expression and morphological features across species to align embryonic development stages between vertebrates. The tool uses ontologies, databases, and user input to generate distance matrices and heat maps showing commonalities. It aims to standardize stage terminology and help comparative biology by identifying conserved developmental processes. Future improvements include using a more detailed anatomical ontology and clustering analyses.