MH prediction modeling and validation in r (1) regression 190709
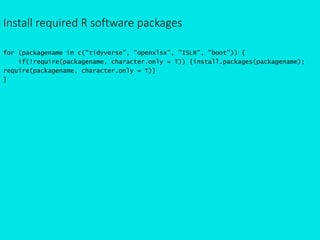
- 1. Install required R software packages for (packagename in c("tidyverse", "openxlsx", "ISLR", "boot")) { if(!require(packagename, character.only = T)) {install.packages(packagename); require(packagename, character.only = T)} }
- 2. Dataset for practice # Assign the data to an object named "dataset" # Save the original row numbers as a separate column~! dataset = ISLR::Auto %>% {add_column(., rownum = 1:nrow(.), .before = 1)} %>% as_data_frame dataset # # A tibble: 392 x 10 # rownum mpg cylinders displacement horsepower weight acceleration year origin name # <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> # 1 1 18 8 307 130 3504 12 70 1 chevrolet chevelle malibu # 2 2 15 8 350 165 3693 11.5 70 1 buick skylark 320 # 3 3 18 8 318 150 3436 11 70 1 plymouth satellite # 4 4 16 8 304 150 3433 12 70 1 amc rebel sst # 5 5 17 8 302 140 3449 10.5 70 1 ford torino # 6 6 15 8 429 198 4341 10 70 1 ford galaxie 500 # 7 7 14 8 454 220 4354 9 70 1 chevrolet impala # 8 8 14 8 440 215 4312 8.5 70 1 plymouth fury iii # 9 9 14 8 455 225 4425 10 70 1 pontiac catalina # 10 10 15 8 390 190 3850 8.5 70 1 amc ambassador dpl # # ... with 382 more rows
- 3. Dataset for practice dataset %>% str # Classes ‘tbl_df’, ‘tbl’ and 'data.frame': 392 obs. of 10 variables: # $ rownum : int 1 2 3 4 5 6 7 8 9 10 ... # $ mpg : num 18 15 18 16 17 15 14 14 14 15 ... # $ cylinders : num 8 8 8 8 8 8 8 8 8 8 ... # $ displacement: num 307 350 318 304 302 429 454 440 455 390 ... # $ horsepower : num 130 165 150 150 140 198 220 215 225 190 ... # $ weight : num 3504 3693 3436 3433 3449 ... # $ acceleration: num 12 11.5 11 12 10.5 10 9 8.5 10 8.5 ... # $ year : num 70 70 70 70 70 70 70 70 70 70 ... # $ origin : num 1 1 1 1 1 1 1 1 1 1 ... # $ name : Factor w/ 304 levels "amc ambassador brougham",..: 49 36 231 14 161 141 54 223 241 2 ...
- 4. Auto {ISLR} R Documentation Auto Data Set Description Gas mileage, horsepower, and other information for 392 vehicles. Format A data frame with 392 observations on the following 9 variables. mpg miles per gallon cylinders Number of cylinders between 4 and 8 displacement Engine displacement (cu. inches) horsepower Engine horsepower weight Vehicle weight (lbs.) acceleration Time to accelerate from 0 to 60 mph (sec.) year Model year (modulo 100) origin Origin of car (1. American, 2. European, 3. Japanese) name Vehicle name
- 6. "Random" sampling to divide training & test data set.seed(1); rownumber4train = sample(392,196) rownumber4train %>% dput # c(105L, 146L, 224L, 354L, 79L, 348L, 365L, 255L, 242L, 24L, 388L, # 68L, 262L, 391L, 292L, 188L, 270L, 372L, 143L, 290L, 387L, 382L, # 384L, 47L, 99L, 142L, 5L, 140L, 317L, 124L, 175L, 217L, 178L, # 67L, 297L, 239L, 283L, 39L, 257L, 379L, 289L, 228L, 275L, 194L, # 185L, 274L, 9L, 165L, 252L, 238L, 164L, 294L, 149L, 83L, 383L, # 34L, 107L, 174L, 222L, 136L, 304L, 98L, 152L, 110L, 214L, 85L, # 157L, 250L, 28L, 356L, 329L, 376L, 111L, 336L, 330L, 323L, 347L, # 123L, 245L, 301L, 333L, 334L, 363L, 101L, 234L, 63L, 218L, 38L, # 75L, 44L, 73L, 18L, 193L, 263L, 233L, 237L, 135L, 121L, 357L, # 360L, 192L, 103L, 371L, 287L, 183L, 62L, 305L, 137L, 299L, 170L, # 276L, 206L, 100L, 295L, 42L, 4L, 198L, 29L, 315L, 362L, 321L, # 296L, 131L, 369L, 203L, 122L, 312L, 55L, 61L, 326L, 151L, 21L, # 10L, 167L, 240L, 154L, 144L, 271L, 251L, 129L, 173L, 380L, 60L, # 65L, 181L, 112L, 303L, 288L, 26L, 211L, 340L, 385L, 373L, 109L, # 120L, 43L, 125L, 313L, 249L, 50L, 359L, 207L, 291L, 179L, 201L, # 94L, 15L, 76L, 163L, 225L, 386L, 186L, 189L, 86L, 339L, 195L, # 311L, 160L, 130L, 300L, 307L, 41L, 187L, 106L, 314L, 40L, 284L, # 370L, 213L, 247L, 256L, 258L, 261L, 375L, 57L, 117L)
- 7. "Random" sampling to divide training & test data dataset.train = dataset[rownumber4train, ] dataset.test = dataset[-rownumber4train, ] dataset.train # # A tibble: 196 x 10 # rownum mpg cylinders displacement horsepower weight acceleration year origin name # <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> # 1 105 13 8 360 170 4654 13 73 1 plymouth custom suburb # 2 146 24 4 90 75 2108 15.5 74 2 fiat 128 # 3 224 17.5 6 250 110 3520 16.4 77 1 chevrolet concours # 4 354 31.6 4 120 74 2635 18.3 81 3 mazda 626 # 5 79 26 4 96 69 2189 18 72 2 renault 12 (sw) # 6 348 34.4 4 98 65 2045 16.2 81 1 ford escort 4w # 7 365 34 4 112 88 2395 18 82 1 chevrolet cavalier 2-door # 8 255 20.5 6 225 100 3430 17.2 78 1 plymouth volare # 9 242 21.5 3 80 110 2720 13.5 77 3 mazda rx-4 # 10 24 26 4 121 113 2234 12.5 70 2 bmw 2002 # # ... with 186 more rows dataset.test # # A tibble: 196 x 10 # rownum mpg cylinders displacement horsepower weight acceleration year origin name # <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> # 1 1 18 8 307 130 3504 12 70 1 chevrolet chevelle malibu # 2 2 15 8 350 165 3693 11.5 70 1 buick skylark 320 # 3 3 18 8 318 150 3436 11 70 1 plymouth satellite # 4 6 15 8 429 198 4341 10 70 1 ford galaxie 500 # 5 7 14 8 454 220 4354 9 70 1 chevrolet impala # 6 8 14 8 440 215 4312 8.5 70 1 plymouth fury iii # 7 11 15 8 383 170 3563 10 70 1 dodge challenger se # 8 12 14 8 340 160 3609 8 70 1 plymouth 'cuda 340 # 9 13 15 8 400 150 3761 9.5 70 1 chevrolet monte carlo # 10 14 14 8 455 225 3086 10 70 1 buick estate wagon (sw) # # ... with 186 more rows
- 8. Check if the train dataset and the test dataset add up to the original dataset #@ Check if the train dataset and the test dataset add up to the original dataset. ---- bind_rows(dataset.train, dataset.test) %>% arrange(rownum) # # A tibble: 392 x 10 # rownum mpg cylinders displacement horsepower weight acceleration year # <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 1 18 8 307 130 3504 12 70 # 2 2 15 8 350 165 3693 11.5 70 # 3 3 18 8 318 150 3436 11 70 # 4 4 16 8 304 150 3433 12 70 # 5 5 17 8 302 140 3449 10.5 70 # 6 6 15 8 429 198 4341 10 70 # 7 7 14 8 454 220 4354 9 70 # 8 8 14 8 440 215 4312 8.5 70 # 9 9 14 8 455 225 4425 10 70 # 10 10 15 8 390 190 3850 8.5 70 # # ... with 382 more rows, and 2 more variables: origin <dbl>, name <fct>
- 9. Check if the train dataset and the test dataset add up to the original dataset all.equal(dataset, bind_rows(dataset.train, dataset.test)) # TRUE
- 10. Save and check the splitted train dataset and test dataset Remove the test dataset until we finish the modeling~! #@ Save and check the splitted train dataset and test dataset. ---- saveRDS(dataset.train, "dataset.train.rds") saveRDS(dataset.test, "dataset.test.rds") #@ You may also export to MS Excel format to check the data. ---- write.xlsx(dataset.train, "dataset.train.xlsx", asTable = T) write.xlsx(dataset.test, "dataset.test.xlsx", asTable = T) openXL("dataset.train.xlsx") openXL("dataset.test.xlsx") #@ Remove the original dataset and the test dataset (to make it unseen until the model is fitted). ---- rm(dataset) rm(dataset.test)
- 12. Visualize mpg vs. horsepower #@ Visualize mpg vs. horsepower ---- # Tools -> Global Options -> R Markdown -> Show output inline for all R Markdown documents # Tools -> Global Options -> R Markdown -> "Show output preview in" -> select "Viewer Pane" dataset.train %>% ggplot(aes(x = horsepower, y = mpg)) + geom_point()
- 13. Visualize mpg vs. horsepower dataset.train %>% ggplot(aes(x = horsepower, y = mpg)) + geom_point() + geom_smooth(method = "lm")
- 14. Regress mpg vs. horsepower #@ Regress mpg vs. horsepower ---- model1 = lm(mpg ~ horsepower, data = dataset.train) model1 # Call: # lm(formula = mpg ~ horsepower, data = dataset.train) # # Coefficients: # (Intercept) horsepower # 40.3404 -0.1617
- 15. Regress mpg vs. horsepower model1 %>% str # List of 12 # $ coefficients : Named num [1:2] 40.34 -0.162 # ..- attr(*, "names")= chr [1:2] "(Intercept)" "horsepower" # $ residuals : Named num [1:196] 0.149 -4.213 -5.053 3.226 -3.183 ... # ..- attr(*, "names")= chr [1:196] "1" "2" "3" "4" ... # $ effects : Named num [1:196] -322.03 -86.12 -5.05 3.49 -2.88 ... # ..- attr(*, "names")= chr [1:196] "(Intercept)" "horsepower" "" "" ... # $ rank : int 2 # $ fitted.values: Named num [1:196] 12.9 28.2 22.6 28.4 29.2 ... # ..- attr(*, "names")= chr [1:196] "1" "2" "3" "4" ... # $ assign : int [1:2] 0 1 # $ qr :List of 5 # ..$ qr : num [1:196, 1:2] -14 0.0714 0.0714 0.0714 0.0714 ... # .. ..- attr(*, "dimnames")=List of 2 # .. .. ..$ : chr [1:196] "1" "2" "3" "4" ... # .. .. ..$ : chr [1:2] "(Intercept)" "horsepower" # .. ..- attr(*, "assign")= int [1:2] 0 1 # ..$ qraux: num [1:2] 1.07 1.07 # ..$ pivot: int [1:2] 1 2 # ..$ tol : num 1e-07 # ..$ rank : int 2 # ..- attr(*, "class")= chr "qr" # $ df.residual : int 194 # $ xlevels : Named list() # $ call : language lm(formula = mpg ~ horsepower, data = dataset.train) # $ terms :Classes 'terms', 'formula' language mpg ~ horsepower # .. ..- attr(*, "variables")= language list(mpg, horsepower) # .. ..- attr(*, "factors")= int [1:2, 1] 0 1 # .. .. ..- attr(*, "dimnames")=List of 2 # .. .. .. ..$ : chr [1:2] "mpg" "horsepower" # .. .. .. ..$ : chr "horsepower" # .. ..- attr(*, "term.labels")= chr "horsepower" # .. ..- attr(*, "order")= int 1 # .. ..- attr(*, "intercept")= int 1 # .. ..- attr(*, "response")= int 1 # .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv> # .. ..- attr(*, "predvars")= language list(mpg, horsepower) # .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric" # .. .. ..- attr(*, "names")= chr [1:2] "mpg" "horsepower" # $ model :'data.frame': 196 obs. of 2 variables: # ..$ mpg : num [1:196] 13 24 17.5 31.6 26 34.4 34 20.5 21.5 26 ... # ..$ horsepower: num [1:196] 170 75 110 74 69 65 88 100 110 113 ... # ..- attr(*, "terms")=Classes 'terms', 'formula' language mpg ~ horsepower # .. .. ..- attr(*, "variables")= language list(mpg, horsepower) # .. .. ..- attr(*, "factors")= int [1:2, 1] 0 1 # .. .. .. ..- attr(*, "dimnames")=List of 2 # .. .. .. .. ..$ : chr [1:2] "mpg" "horsepower" # .. .. .. .. ..$ : chr "horsepower" # .. .. ..- attr(*, "term.labels")= chr "horsepower" # .. .. ..- attr(*, "order")= int 1 # .. .. ..- attr(*, "intercept")= int 1 # .. .. ..- attr(*, "response")= int 1 # .. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv> # .. .. ..- attr(*, "predvars")= language list(mpg, horsepower) # .. .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric" # .. .. .. ..- attr(*, "names")= chr [1:2] "mpg" "horsepower" # - attr(*, "class")= chr "lm"
- 16. Regress mpg vs. horsepower model1$coefficients %>% dput # c(`(Intercept)` = 40.3403771907169, horsepower = -0.161701271858609) model1$coefficients[1] # (Intercept) # 40.34038 model1$coefficients[2] # horsepower # -0.1617013
- 17. Calculate the training mean squared error dataset.train = dataset.train %>% mutate(mpg.tmp.predict = model1$coefficients[1] + model1$coefficients[2] * horsepower) dataset.train = dataset.train %>% mutate(mpg.tmp.residual = model1$coefficients[1] + model1$coefficients[2] * horsepower - mpg) dataset.train = dataset.train %>% mutate(mpg.tmp2.predict = 50 - 0.3 * horsepower) dataset.train = dataset.train %>% mutate(mpg.tmp2.residual = 50 - 0.3 * horsepower - mpg) dataset.train %>% select(horsepower, mpg, mpg.tmp.predict, mpg.tmp.residual, mpg.tmp2.predict, mpg.tmp2.residual) %>% arrange(horsepower) # # A tibble: 196 x 6 # horsepower mpg mpg.tmp.predict mpg.tmp.residual mpg.tmp2.predict mpg.tmp2.residual # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 49 29 32.4 3.42 35.3 6.30 # 2 52 31 31.9 0.932 34.4 3.40 # 3 52 29 31.9 2.93 34.4 5.40 # 4 52 32.8 31.9 -0.868 34.4 1.6 # 5 58 36 31.0 -5.04 32.6 -3.40 # 6 58 39.1 31.0 -8.14 32.6 -6.5 # 7 60 27 30.6 3.64 32 5 # 8 60 24.5 30.6 6.14 32 7.5 # 9 60 36.1 30.6 -5.46 32 -4.1 # 10 61 32 30.5 -1.52 31.7 -0.3 # # ... with 186 more rows
- 18. Calculate the training mean squared error dataset.train %>% ggplot(aes(x = horsepower, y = mpg)) + geom_point() + geom_smooth(method = "lm") + geom_abline(intercept = 50, slope = -0.3, color = "red")
- 19. Calculate the training mean squared error dataset.train$mpg.tmp.residual %>% mean dataset.train$mpg.tmp.residual^2 %>% mean # [1] -3.697654e-15 # [1] 21.78987 dataset.train$mpg.tmp.residual^2 %>% mean dataset.train$mpg.tmp2.residual^2 %>% mean # [1] 21.78987 # [1] 76.1948
- 20. Calculate the training mean squared error dataset.train = dataset.train %>% mutate(mpg.tmp.predict = model1$coefficients[1] + model1$coefficients[2] * horsepower) dataset.train = dataset.train %>% mutate(mpg.tmp.residual = model1$coefficients[1] + model1$coefficients[2] * horsepower - mpg) dataset.train = dataset.train %>% {mutate(., mpg.model1.predict = predict(model1, newdata = .))} dataset.train = dataset.train %>% {mutate(., mpg.model1.residual = predict(model1, newdata = .) - mpg)} dataset.train %>% select(horsepower, mpg, mpg.model1.predict, mpg.tmp.predict, mpg.model1.residual, mpg.tmp.residual) %>% arrange(horsepower) dataset.train$mpg.model1.residual %>% mean dataset.train$mpg.model1.residual^2 %>% mean # # A tibble: 196 x 6 # horsepower mpg mpg.model1.predict mpg.tmp.predict mpg.model1.residual mpg.tmp.residual # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 49 29 32.4 32.4 3.42 3.42 # 2 52 31 31.9 31.9 0.932 0.932 # 3 52 29 31.9 31.9 2.93 2.93 # 4 52 32.8 31.9 31.9 -0.868 -0.868 # 5 58 36 31.0 31.0 -5.04 -5.04 # 6 58 39.1 31.0 31.0 -8.14 -8.14 # 7 60 27 30.6 30.6 3.64 3.64 # 8 60 24.5 30.6 30.6 6.14 6.14 # 9 60 36.1 30.6 30.6 -5.46 -5.46 # 10 61 32 30.5 30.5 -1.52 -1.52 # # ... with 186 more rows # [1] -3.842838e-15 # [1] 21.78987
- 22. Calculate the test mean squared error #@ Loading the test dataset (after modeling is finished~!) ----- dataset.test = readRDS("dataset.test.rds") # Same codes using `$` operator & `%>%` operator. # dataset.test$mpg.model1.predict = predict(model1, newdata = dataset.test) # dataset.test$mpg.model1.residual = predict(model1, newdata = dataset.test) - dataset.test$mpg dataset.test = dataset.test %>% {mutate(., mpg.model1.predict = predict(model1, newdata = .))} dataset.test = dataset.test %>% {mutate(., mpg.model1.residual = predict(model1, newdata = .) - mpg)} dataset.test %>% select(horsepower, mpg, mpg.model1.predict, mpg.model1.residual) %>% arrange(horsepower) # # A tibble: 196 x 4 # horsepower mpg mpg.model1.predict mpg.model1.residual # <dbl> <dbl> <dbl> <dbl> # 1 46 26 32.9 6.90 # 2 46 26 32.9 6.90 # 3 48 43.1 32.6 -10.5 # 4 48 44.3 32.6 -11.7 # 5 48 43.4 32.6 -10.8 # 6 52 44 31.9 -12.1 # 7 53 33 31.8 -1.23 # 8 53 33 31.8 -1.23 # 9 54 23 31.6 8.61 # 10 60 38.1 30.6 -7.46 # # ... with 186 more rows dataset.test$mpg.model1.residual %>% mean # [1] 0.003251906 dataset.test$mpg.model1.residual^2 %>% mean # [1] 26.14142 #@ Remove the test dataset (before any additional modeling~!) ----- rm(dataset.test)
- 23. Visualize mpg vs. horsepower polynomial #@ Visualize mpg vs. horsepower polynomial ---- dataset.train %>% ggplot(aes(x = horsepower, y = mpg)) + geom_point() + geom_smooth(method = "lm") + stat_smooth(method="lm", formula = y ~ poly(x, 2), color="red", fill = "red") + stat_smooth(method="lm", formula = y ~ poly(x, 3), color="orange", fill = "orange")
- 25. Calculate the training MSE & test MSE for multiple models #@ Regress mpg vs. horsepower: simplified ---- model1 = glm(mpg ~ horsepower, data = dataset.train) model1$coefficients %>% dput model.MSE = function(model.object, dataset, y) mean((y-predict(model.object, newdata = dataset))^2) model.MSE(model1, dataset.train, dataset.train$mpg) dataset.test = readRDS("dataset.test.rds") model.MSE(model1, dataset.test, dataset.test$mpg) rm(dataset.test) # c(`(Intercept)` = 40.3403771907169, horsepower = -0.161701271858609) # [1] 21.78987 # [1] 26.14142
- 26. Calculate the training MSE & test MSE for multiple models #@ Regress mpg vs. horsepower: simplified ---- model2 = glm(mpg ~ poly(horsepower, 2), data = dataset.train) model2$coefficients %>% dput model.MSE = function(model.object, dataset, y) mean((y-predict(model.object, newdata = dataset))^2) model.MSE(model2, dataset.train, dataset.train$mpg) dataset.test = readRDS("dataset.test.rds") model.MSE(model2, dataset.test, dataset.test$mpg) rm(dataset.test) # c(`(Intercept)` = 23.0020408163265, `poly(horsepower, 2)1` = -86.1241258368344, `poly(horsepower, 2)2` = 26.1867921933764) # [1] 18.29115 # [1] 19.82259
- 27. Calculate the training MSE & test MSE for multiple models #@ Regress mpg vs. horsepower: simplified ---- model3 = glm(mpg ~ poly(horsepower, 3), data = dataset.train) model3$coefficients %>% dput model.MSE = function(model.object, dataset, y) mean((y-predict(model.object, newdata = dataset))^2) model.MSE(model3, dataset.train, dataset.train$mpg) dataset.test = readRDS("dataset.test.rds") model.MSE(model3, dataset.test, dataset.test$mpg) rm(dataset.test) # c(`(Intercept)` = 23.0020408163265, `poly(horsepower, 3)1` = -86.1241258368344, `poly(horsepower, 3)2` = 26.1867921933764, `poly(horsepower, 3)3` = -1.78925693987765) # [1] 18.27482 # [1] 19.78252
- 29. Fit multiple models using for-loop #@ Fit multiple models using for-loop, and then save the models as R list of objects. ===== model.list = list() dataset.train = readRDS("dataset.train.rds") dataset = as.data.frame(dataset.train) for (i in 1:5) { myformula = as.formula(paste0("mpg ~ poly(horsepower, ", i, ")")) model.list[[i]] = glm(myformula, data = dataset) }
- 30. Calculate the training MSE & test MSE for multiple models #@ Loading the test dataset (after modeling is finished~!) ----- dataset.test = readRDS("dataset.test.rds") #@ Define the loss function (optimization objective). ----- # Cf) You may define any function to avoid repetitive codes. MSE = function(y,yhat) mean((y-yhat)^2) # Make a table that shows the training error and test error for each model in the model.list. ----- df = data.frame( i = 1:length(model.list) , trainMSE = model.list %>% map_dbl(function(object) MSE(object$y, predict(object))) , testMSE = model.list %>% map_dbl(function(object) MSE(dataset.test$mpg, predict(object, newdata = dataset.test))) ) df # i trainMSE testMSE # 1 1 21.78987 26.14142 # 2 2 18.29115 19.82259 # 3 3 18.27482 19.78252 # 4 4 18.12455 19.99969 # 5 5 17.45436 20.18225 #@ Remove the test dataset (before any additional modeling~!) ----- rm(dataset.test)
- 31. Calculate the training MSE & test MSE for multiple models df.gather = df %>% gather(key, value, trainMSE, testMSE) df.gather # i key value # 1 1 trainMSE 21.78987 # 2 2 trainMSE 18.29115 # 3 3 trainMSE 18.27482 # 4 4 trainMSE 18.12455 # 5 5 trainMSE 17.45436 # 6 1 testMSE 26.14142 # 7 2 testMSE 19.82259 # 8 3 testMSE 19.78252 # 9 4 testMSE 19.99969 # 10 5 testMSE 20.18225
- 32. Calculate the training MSE & test MSE for multiple models df.gather %>% ggplot(aes(x = i, y = value, color = key)) + geom_point() + geom_line()
- 35. K-fold "random" split of the training dataset #@ K-fold "random" split of the training dataset ----- function.vec.fold.index = function(data, k = 5) data %>% { rep(1:k, (nrow(.) %/% k) + 1) [1:nrow(.)] } dataset.train %>% function.vec.fold.index(k = 5) %>% dput set.seed(12345); dataset.train %>% function.vec.fold.index(k = 5) %>% sample %>% dput # c(1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, # 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, # 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, # 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, # 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, # 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, # 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, # 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, # 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, # 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, # 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, # 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, # 2L, 3L, 4L, 5L, 1L) # c(2L, 1L, 3L, 2L, 3L, 2L, 2L, 2L, 2L, 1L, 2L, 4L, 1L, 1L, 2L, # 4L, 5L, 3L, 1L, 4L, 5L, 3L, 3L, 3L, 1L, 2L, 4L, 2L, 4L, 1L, 2L, # 3L, 1L, 2L, 5L, 4L, 5L, 4L, 3L, 2L, 3L, 5L, 3L, 5L, 5L, 4L, 5L, # 2L, 4L, 2L, 1L, 5L, 1L, 4L, 5L, 1L, 3L, 2L, 1L, 3L, 3L, 5L, 3L, # 1L, 5L, 5L, 4L, 5L, 2L, 1L, 2L, 4L, 2L, 2L, 1L, 5L, 1L, 3L, 3L, # 3L, 4L, 2L, 5L, 3L, 2L, 4L, 1L, 5L, 1L, 4L, 4L, 5L, 4L, 5L, 4L, # 2L, 2L, 2L, 5L, 4L, 1L, 1L, 2L, 5L, 3L, 3L, 3L, 3L, 3L, 4L, 3L, # 3L, 5L, 3L, 1L, 4L, 4L, 1L, 4L, 5L, 3L, 4L, 2L, 5L, 5L, 2L, 5L, # 3L, 4L, 2L, 3L, 4L, 1L, 1L, 5L, 4L, 3L, 1L, 2L, 1L, 5L, 1L, 4L, # 3L, 3L, 1L, 5L, 4L, 5L, 2L, 4L, 1L, 1L, 1L, 4L, 1L, 3L, 3L, 1L, # 2L, 2L, 1L, 1L, 5L, 4L, 1L, 1L, 5L, 4L, 5L, 4L, 5L, 1L, 5L, 5L, # 4L, 5L, 2L, 4L, 3L, 2L, 2L, 5L, 4L, 3L, 2L, 5L, 3L, 2L, 2L, 1L, # 1L, 4L, 3L, 3L, 4L)
- 36. K-fold "random" split of the training dataset #@ K-fold split of the training dataset ----- dataset.train = dataset.train %>% rownames_to_column # Do not forget to set the random seed, before performing any randomization tasks (e.g., random sampling). set.seed(12345); dataset.train$fold.index = dataset.train %>% function.vec.fold.index(k = 5) %>% sample dataset.train %>% select(rownum, mpg, horsepower, fold.index) # # A tibble: 196 x 4 # rownum mpg horsepower fold.index # <int> <dbl> <dbl> <int> # 1 105 13 170 2 # 2 146 24 75 1 # 3 224 17.5 110 3 # 4 354 31.6 74 2 # 5 79 26 69 3 # 6 348 34.4 65 2 # 7 365 34 88 2 # 8 255 20.5 100 2 # 9 242 21.5 110 2 # 10 24 26 113 1 # # ... with 186 more rows
- 37. K-fold "random" split of the training dataset dataset.train %>% select(rownum, mpg, horsepower, fold.index) %>% filter(fold.index != 1) dataset.train %>% select(rownum, mpg, horsepower, fold.index) %>% filter(fold.index == 1) # # A tibble: 156 x 4 # rownum mpg horsepower fold.index # <int> <dbl> <dbl> <int> # 1 105 13 170 2 # 2 224 17.5 110 3 # 3 354 31.6 74 2 # 4 79 26 69 3 # 5 348 34.4 65 2 # 6 365 34 88 2 # 7 255 20.5 100 2 # 8 242 21.5 110 2 # 9 388 27 86 2 # 10 68 13 155 4 # # ... with 146 more rows # # A tibble: 40 x 4 # rownum mpg horsepower fold.index # <int> <dbl> <dbl> <int> # 1 146 24 75 1 # 2 24 26 113 1 # 3 262 17.7 165 1 # 4 391 28 79 1 # 5 143 31 52 1 # 6 99 18 100 1 # 7 124 11 180 1 # 8 178 22 98 1 # 9 164 20 110 1 # 10 149 26 93 1 # # ... with 30 more rows
- 38. K-fold "random" split of the training dataset ## Visual check of the distribution of the folds ---- dataset.train %>% ggplot(aes(x = horsepower, y = mpg, color = as.factor(fold.index))) + geom_point()
- 40. Fit multiple models in each cross-validation folds #@ Nested for-loop: (1) Iteration of folds for cross-validation (2) Fit multiple models using for-loop ===== # Save the models as a "nested" list of objects to save the results from "nested" for-loop. max.polynomial = 10 cv.model.list = list() for (i.fold in sort(unique(dataset.train$fold.index))) { cv.model.list[[i.fold]] = list() dataset = dataset.train %>% filter(fold.index != i.fold) %>% as.data.frame for (i in 1:max.polynomial) { myformula = as.formula(paste0("mpg ~ poly(horsepower, ", i, ")")) cv.model.list[[i.fold]][[i]] = glm(myformula, data = dataset) } }
- 41. Calculate the training MSE & validation MSE for multiple models in each cross-validation folds #@ Define the loss function (optimization objective). ----- # Cf) You may define any function to avoid repetitive codes. MSE = function(y,yhat) mean((y-yhat)^2) # Make a table that shows the training error and test error for each cross-validation & each model in the "nested" model.list. ----- cv.df = data_frame( cv = rep(1:k, each = max.polynomial) , polynomial = rep(1:max.polynomial, k) ) %>% mutate( trainMSE = map2_dbl(cv, polynomial, function(i.fold, i) { cv.model.list[[i.fold]][[i]] %>% {MSE(.$y, predict(.)) } }) , cvMSE = map2_dbl(cv, polynomial, function(i.fold, i) { MSE(dataset.train %>% filter(fold.index == i.fold) %>% select(mpg) %>% unlist, predict(cv.model.list[[i.fold]][[i]], newdata = dataset.train %>% filter(fold.index == i.fold))) } ) ) cv.df # # A tibble: 50 x 4 # cv polynomial trainMSE cvMSE # <int> <int> <dbl> <dbl> # 1 1 1 22.5 19.3 # 2 1 2 18.9 16.3 # 3 1 3 18.9 16.3 # 4 1 4 18.5 17.4 # 5 1 5 17.6 17.4 # 6 1 6 17.4 16.9 # 7 1 7 17.2 16.3 # 8 1 8 17.0 17.9 # 9 1 9 17.0 17.7 # 10 1 10 17.0 17.6 # # ... with 40 more rows
- 42. Calculate the (aggregated) training MSE & (aggregated) cv MSE for multiple models # Make a table that shows the (aggregated) training error and test error for each model ----- cv.df.summarize = cv.df %>% select(-cv) %>% group_by(polynomial) %>% summarize_all(mean) cv.df.summarize # # A tibble: 10 x 3 # polynomial trainMSE cvMSE # <int> <dbl> <dbl> # 1 1 21.7 22.5 # 2 2 18.2 18.8 # 3 3 18.2 18.9 # 4 4 18.0 19.1 # 5 5 17.4 18.2 # 6 6 17.1 18.0 # 7 7 16.7 18.7 # 8 8 16.7 18.8 # 9 9 16.6 19.3 # 10 10 16.6 18.5
- 43. Visualize the (aggregated) training MSE & cv MSE for multiple models cv.df.summarize %>% gather(key, value, trainMSE, cvMSE) %>% ggplot(aes(x = polynomial, y = value, color = key)) + geom_point() + geom_line()