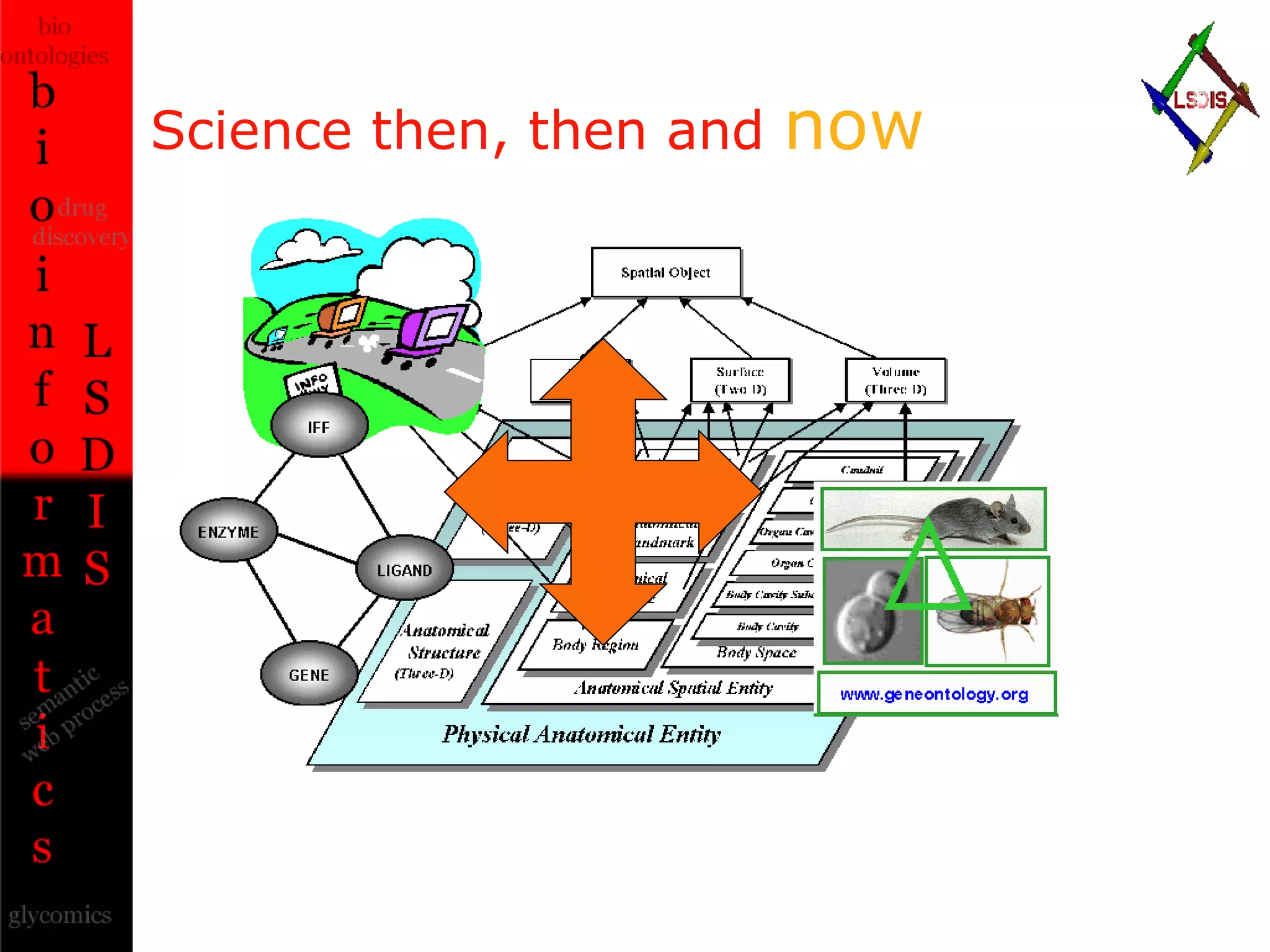
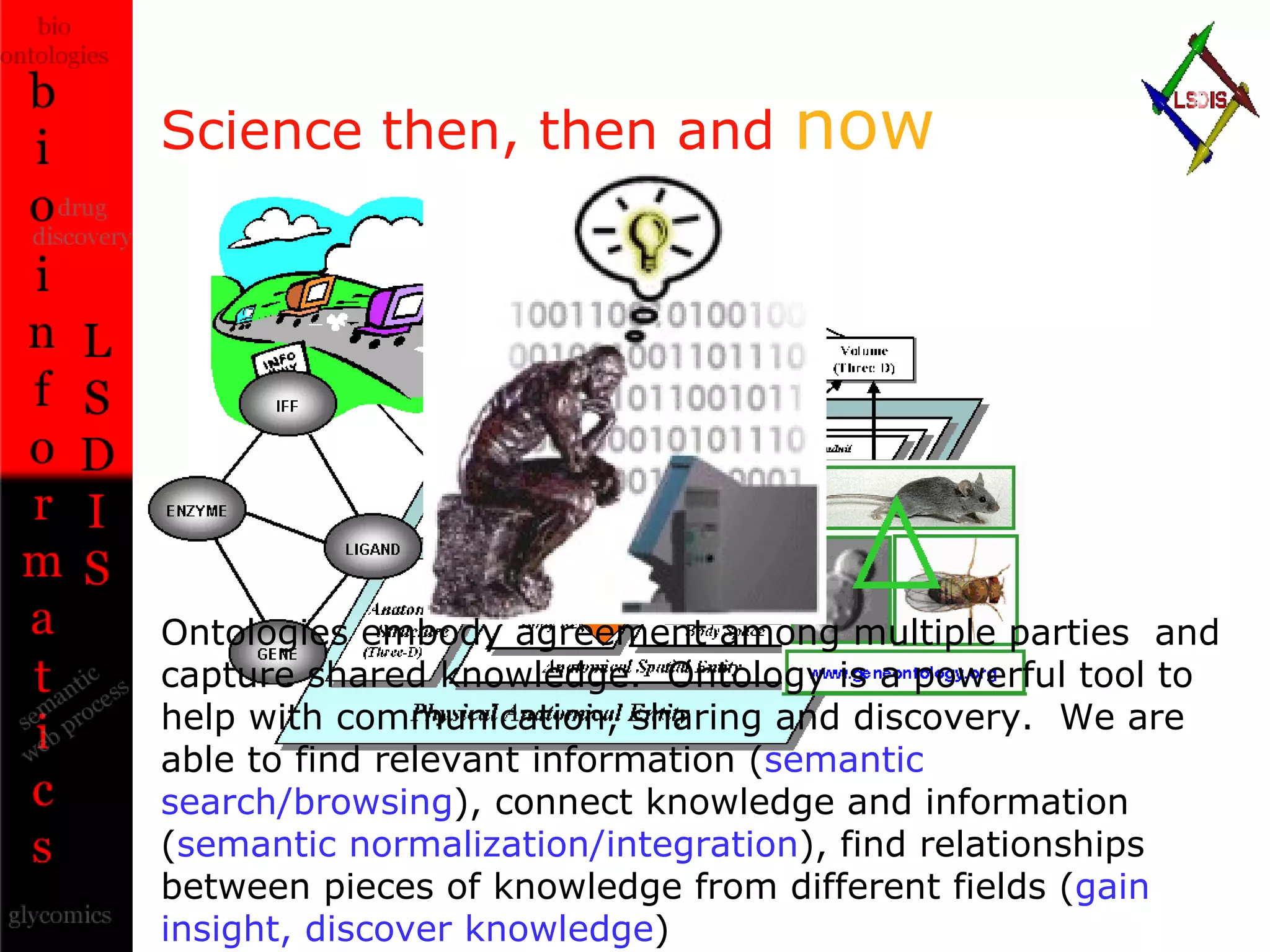
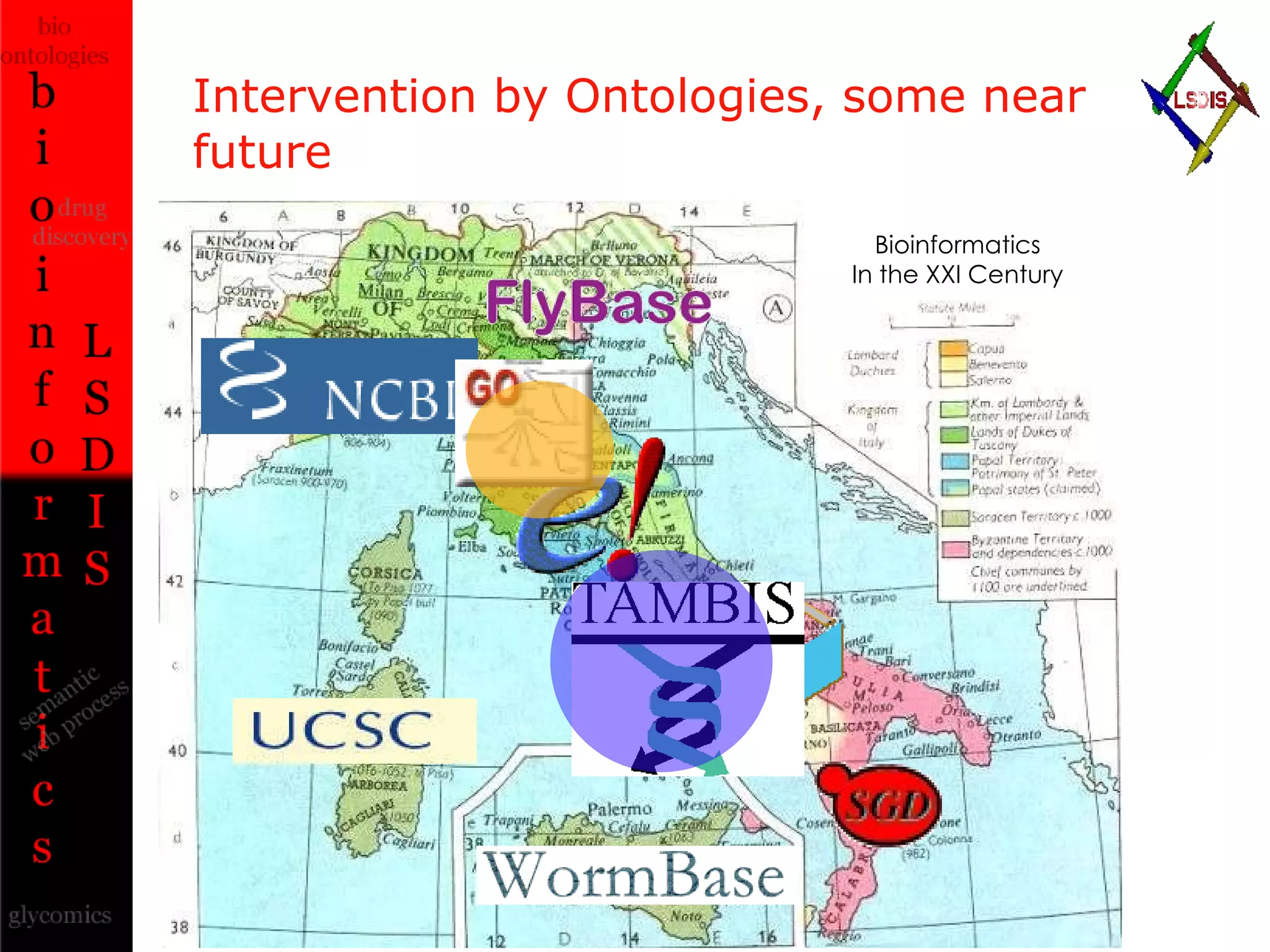
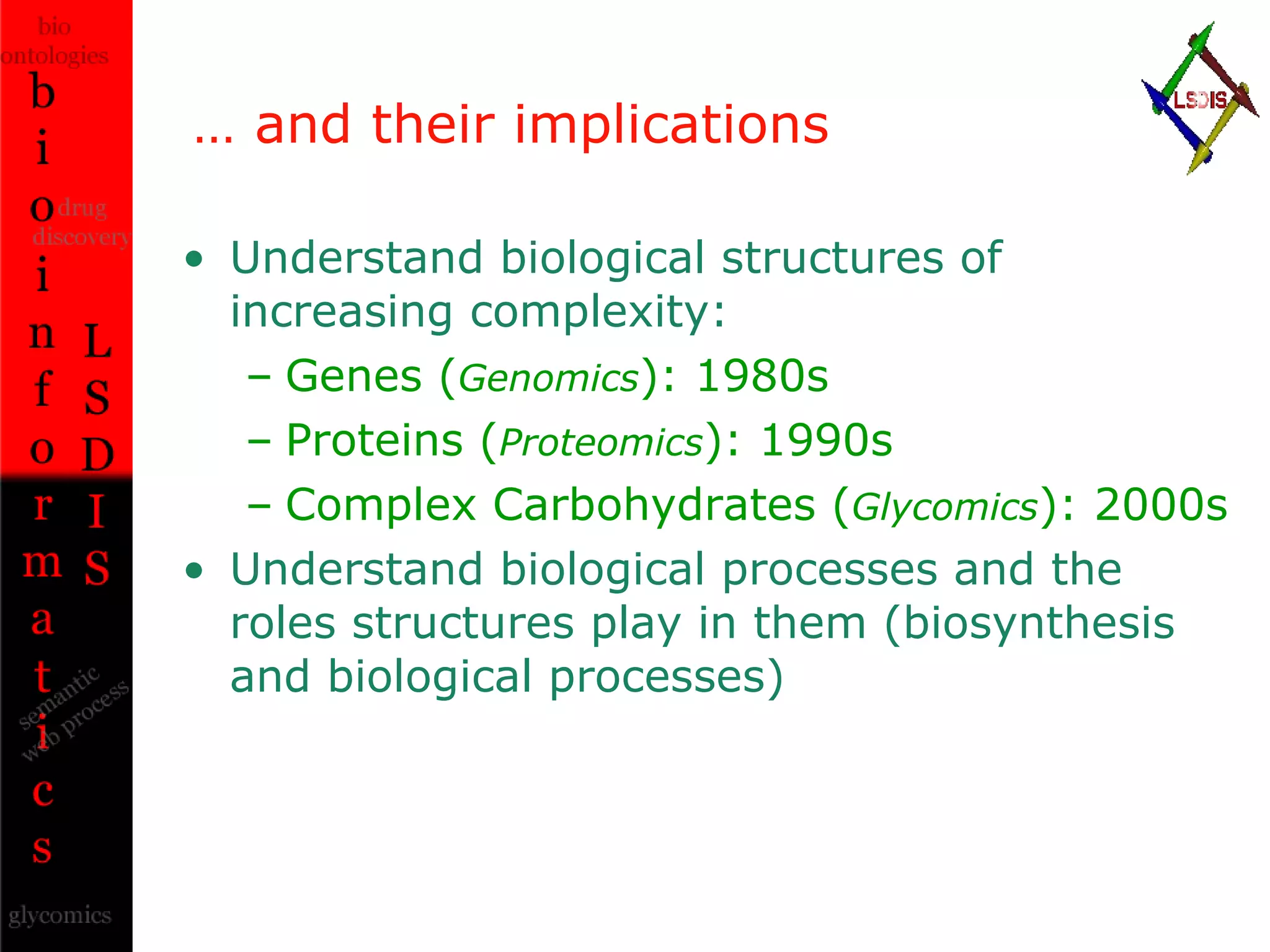
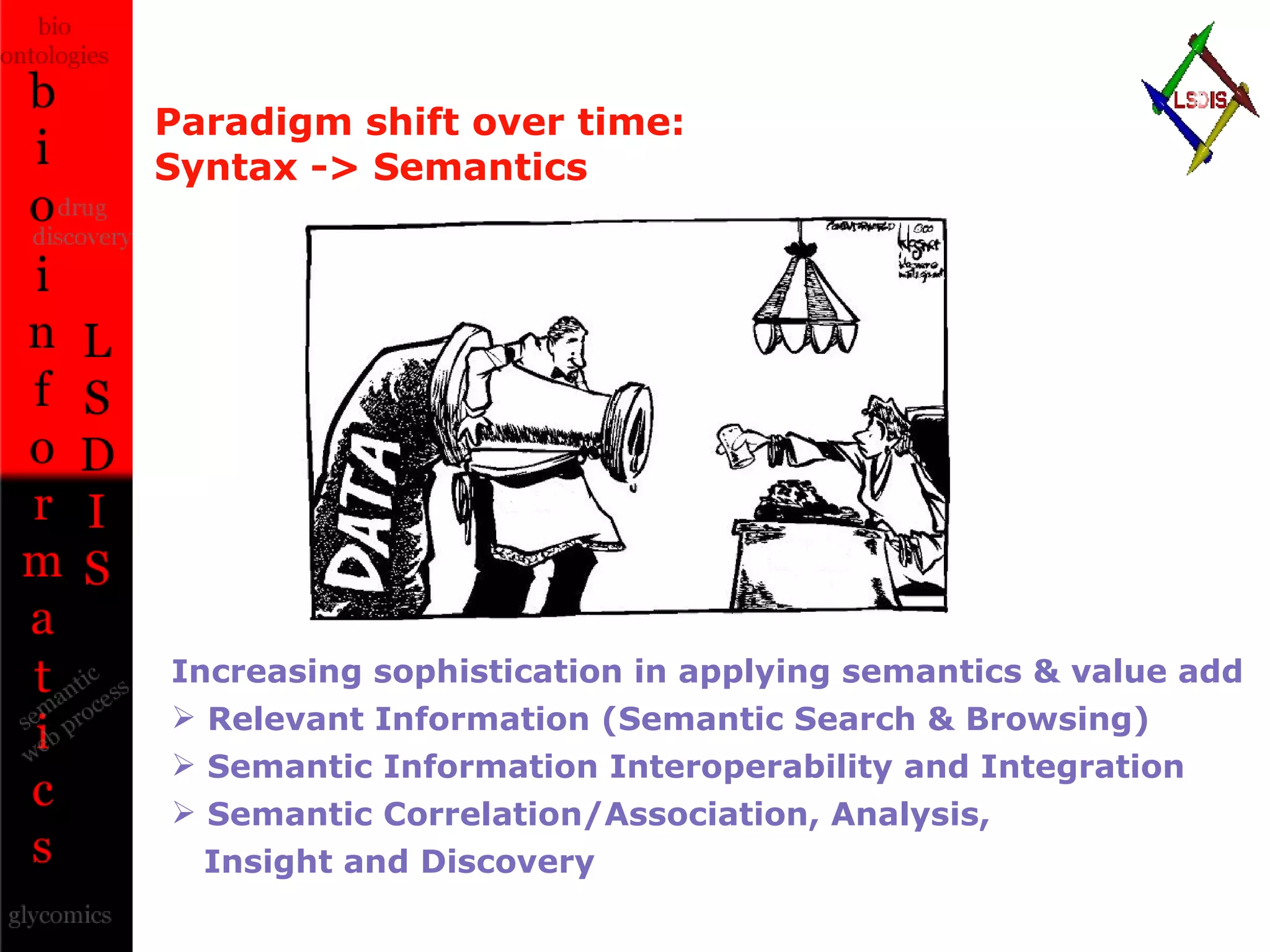
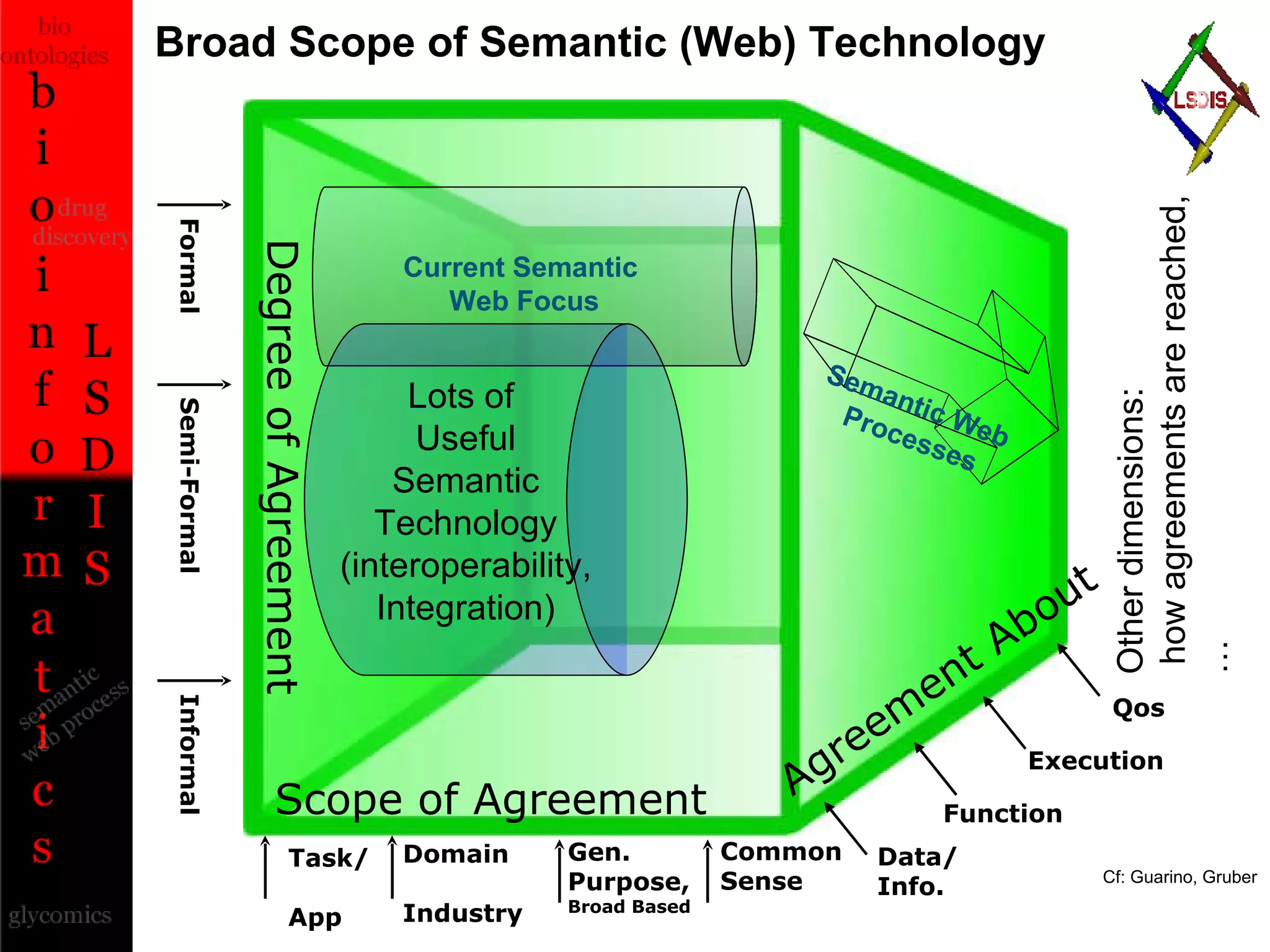
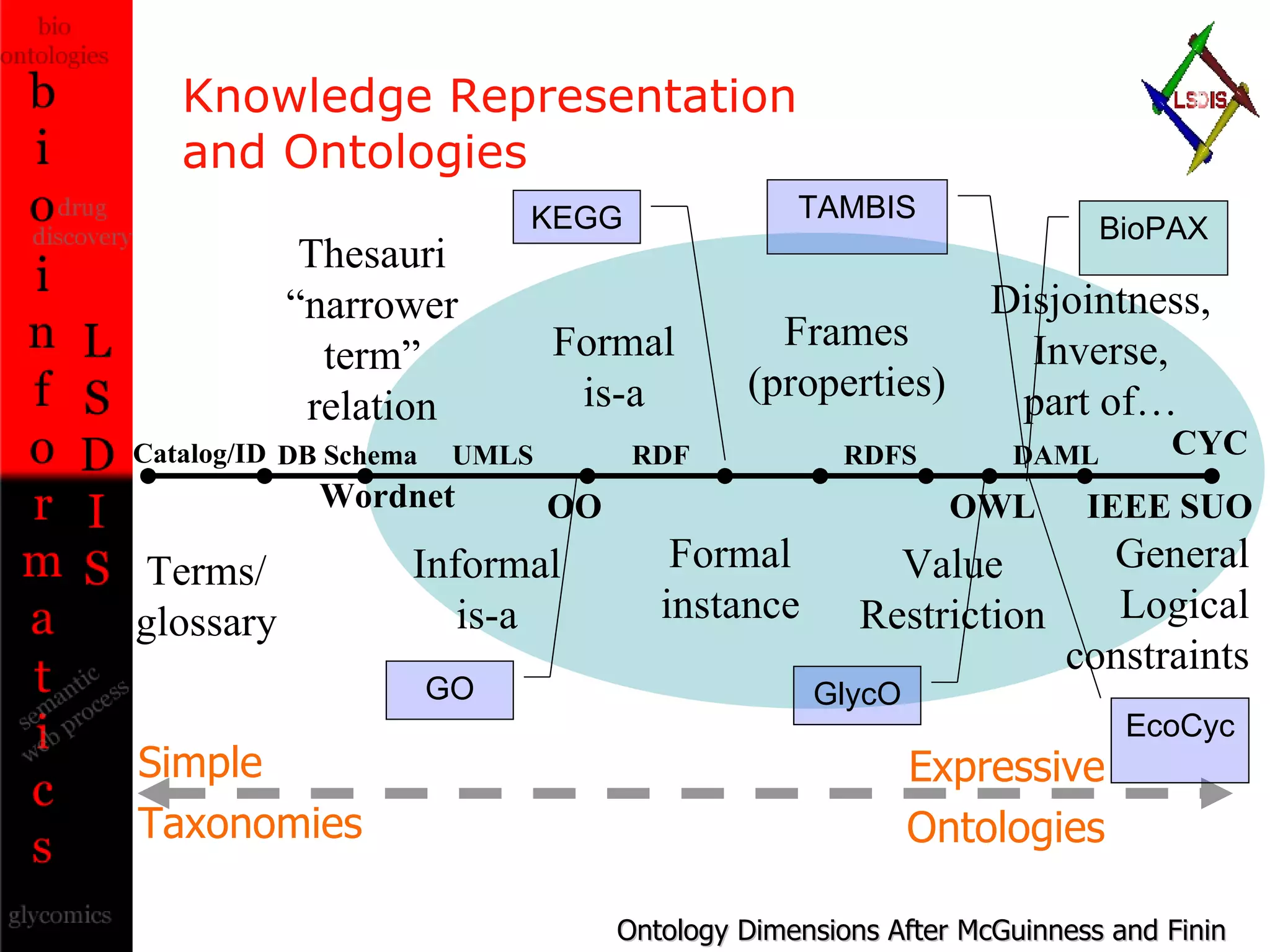
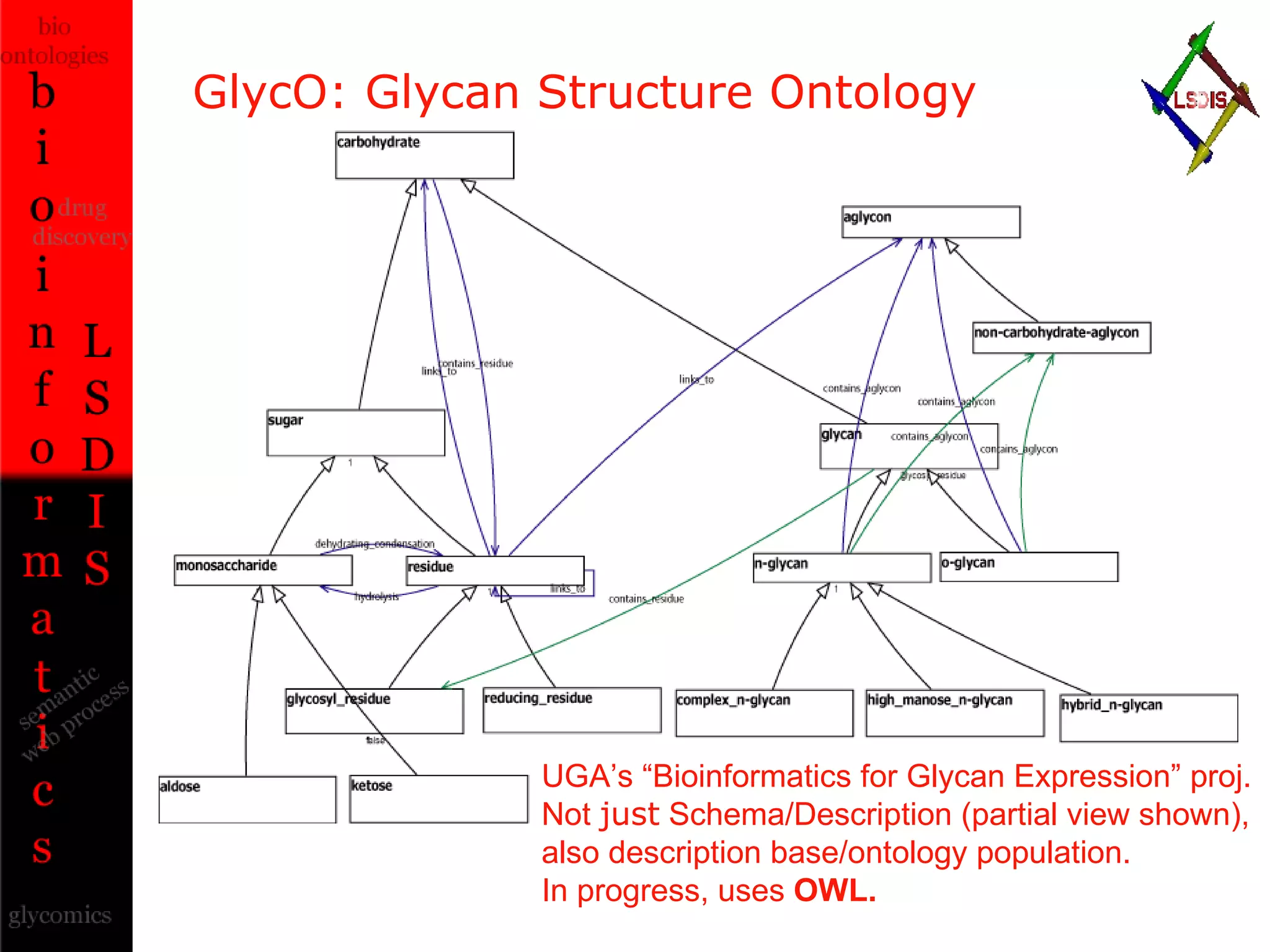
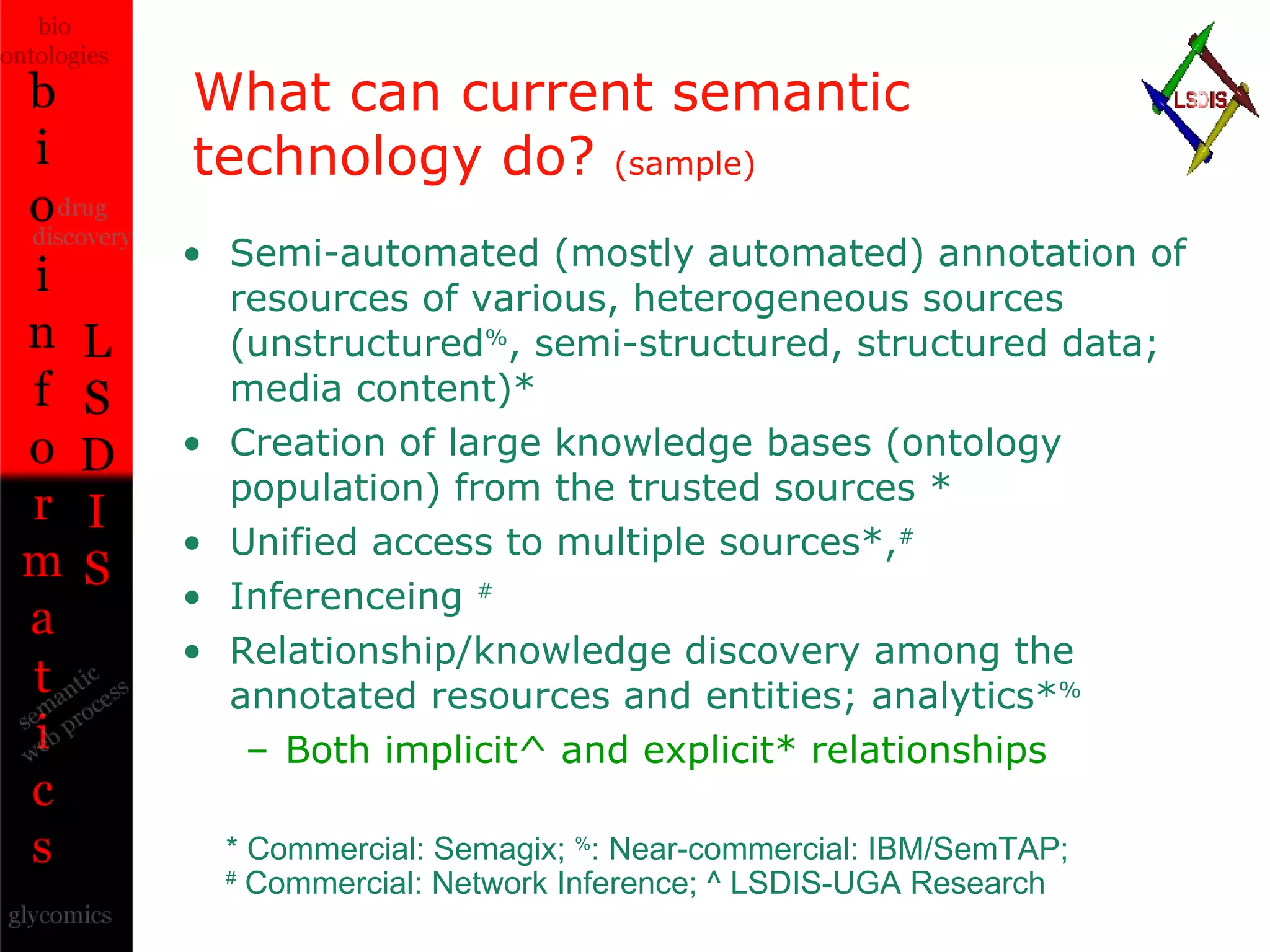
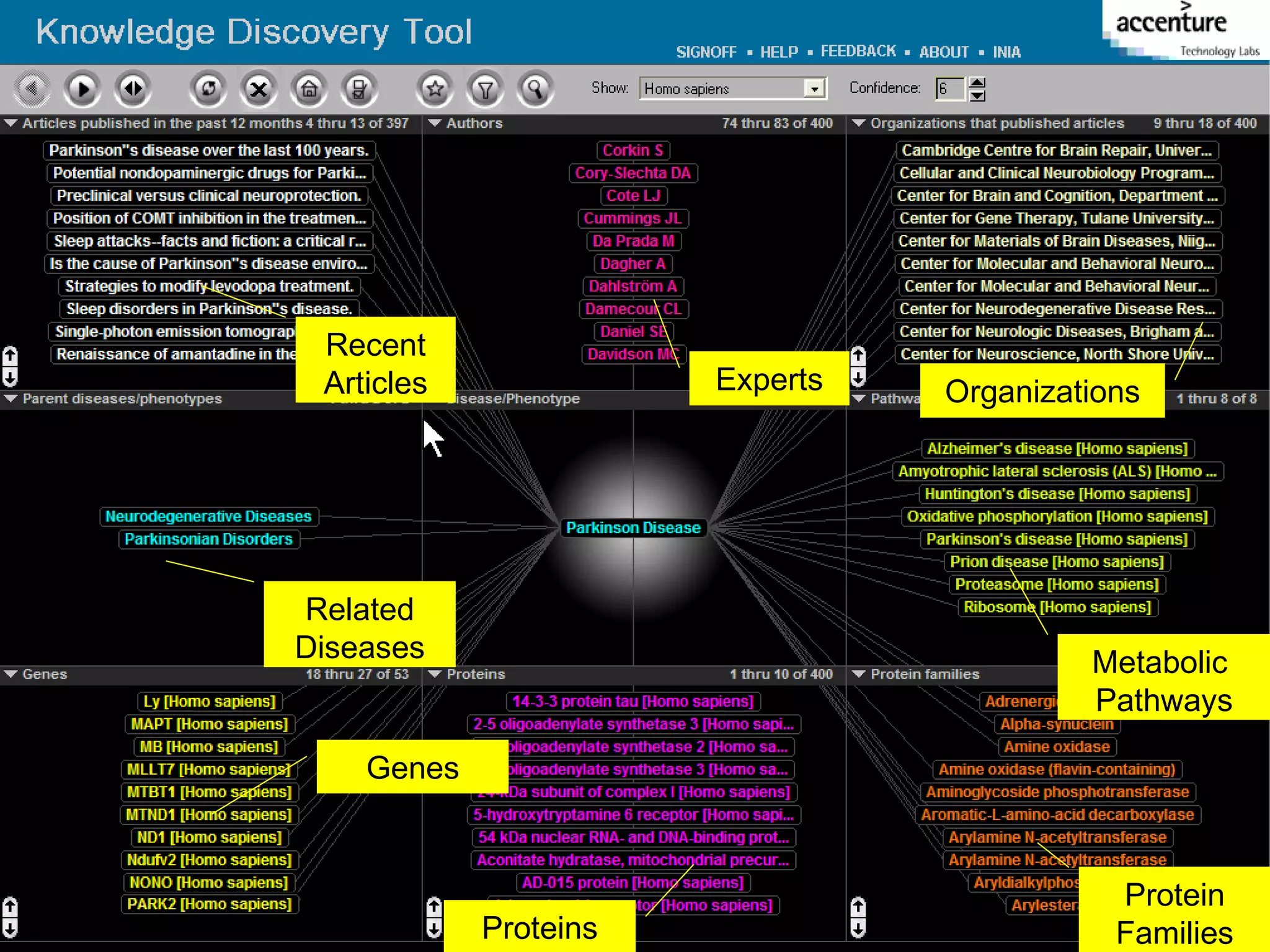
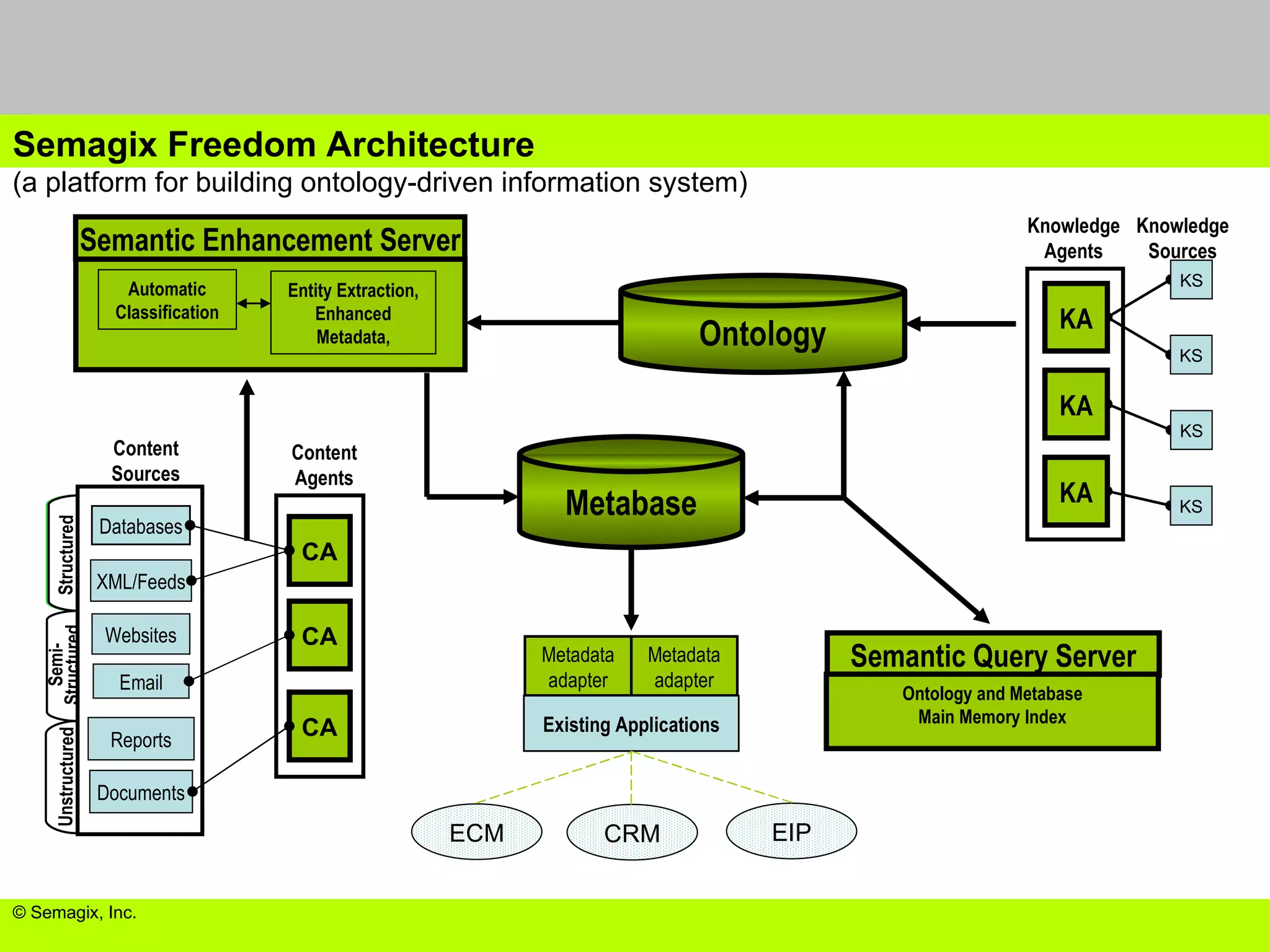
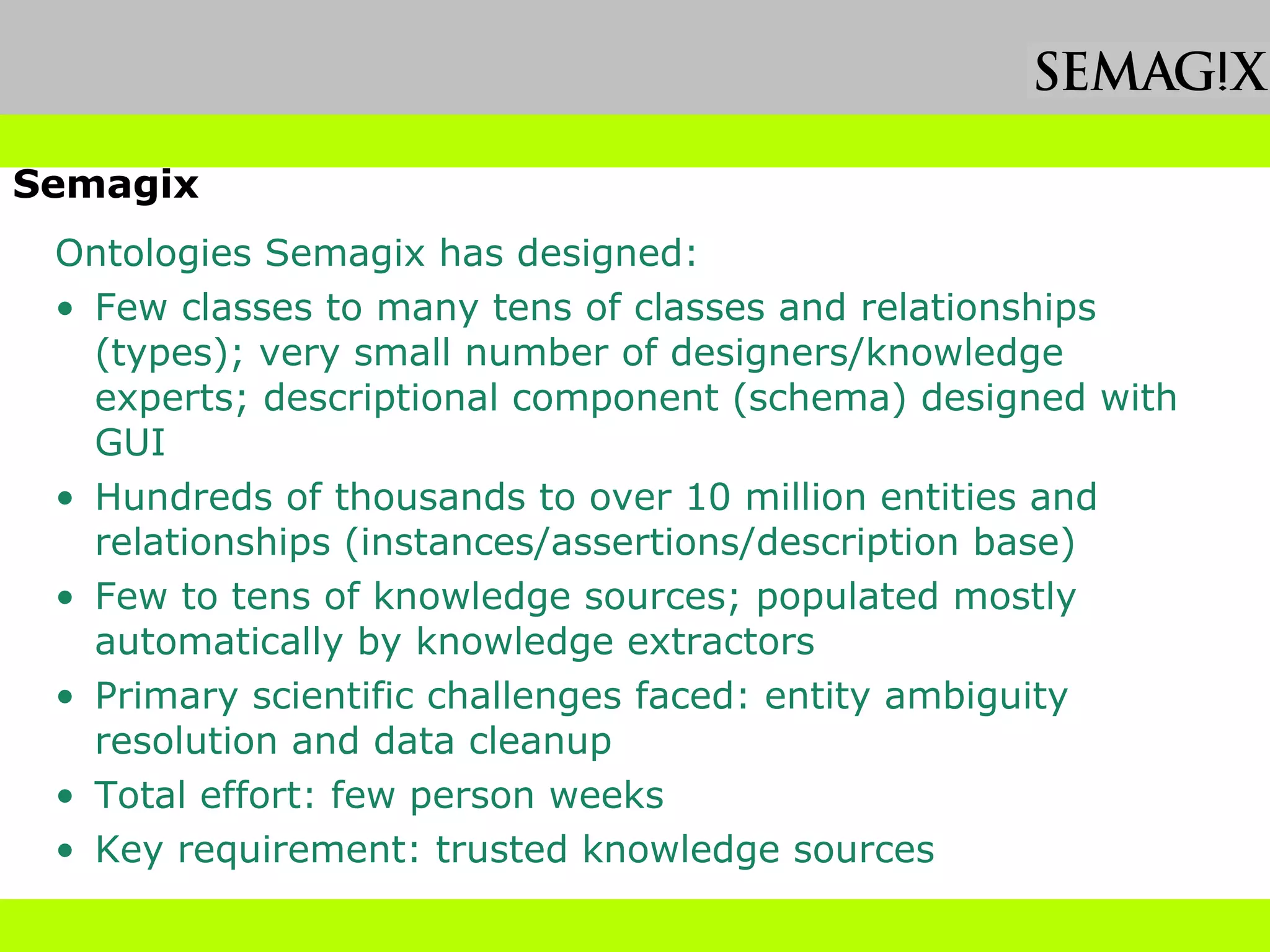
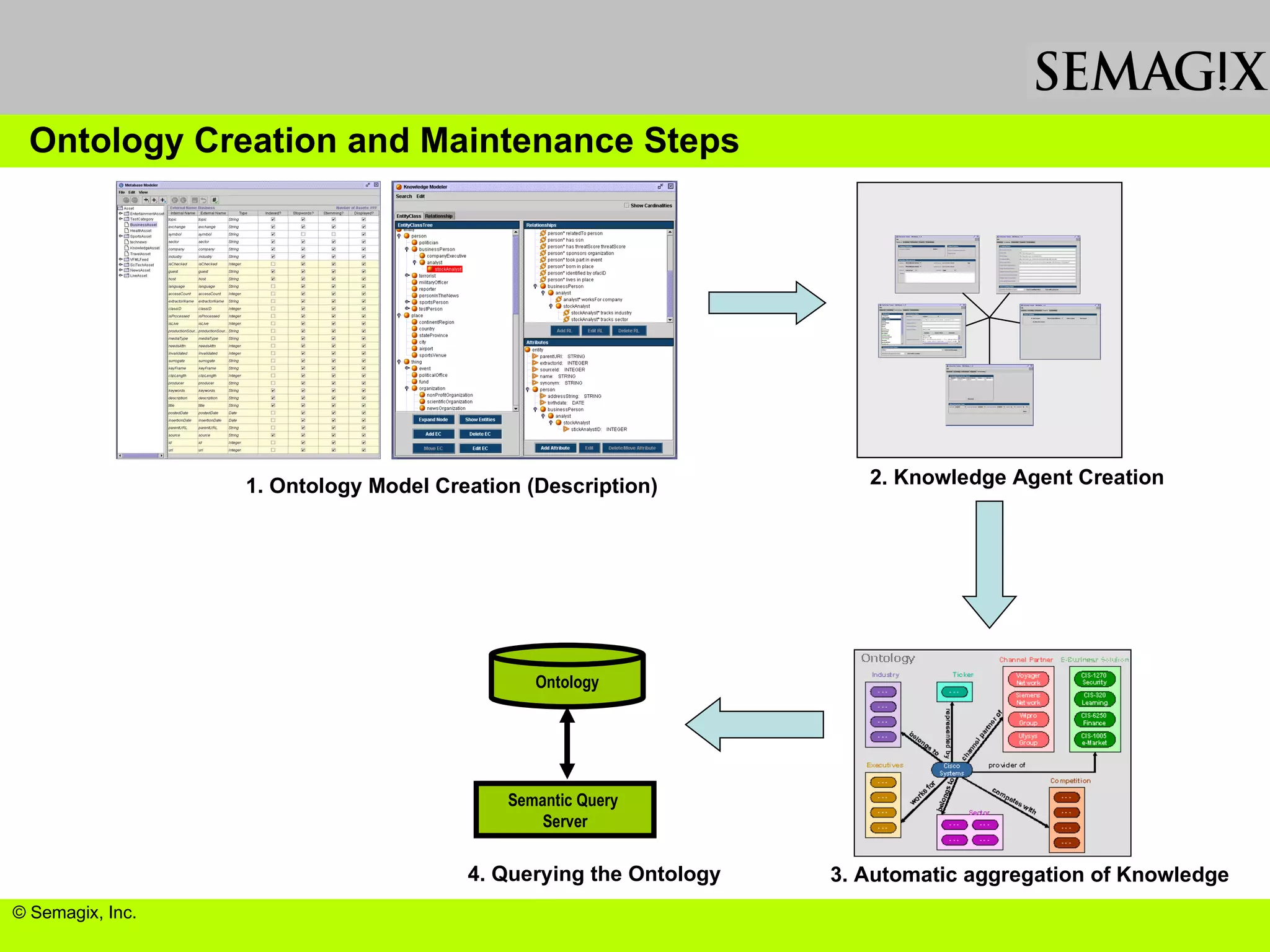
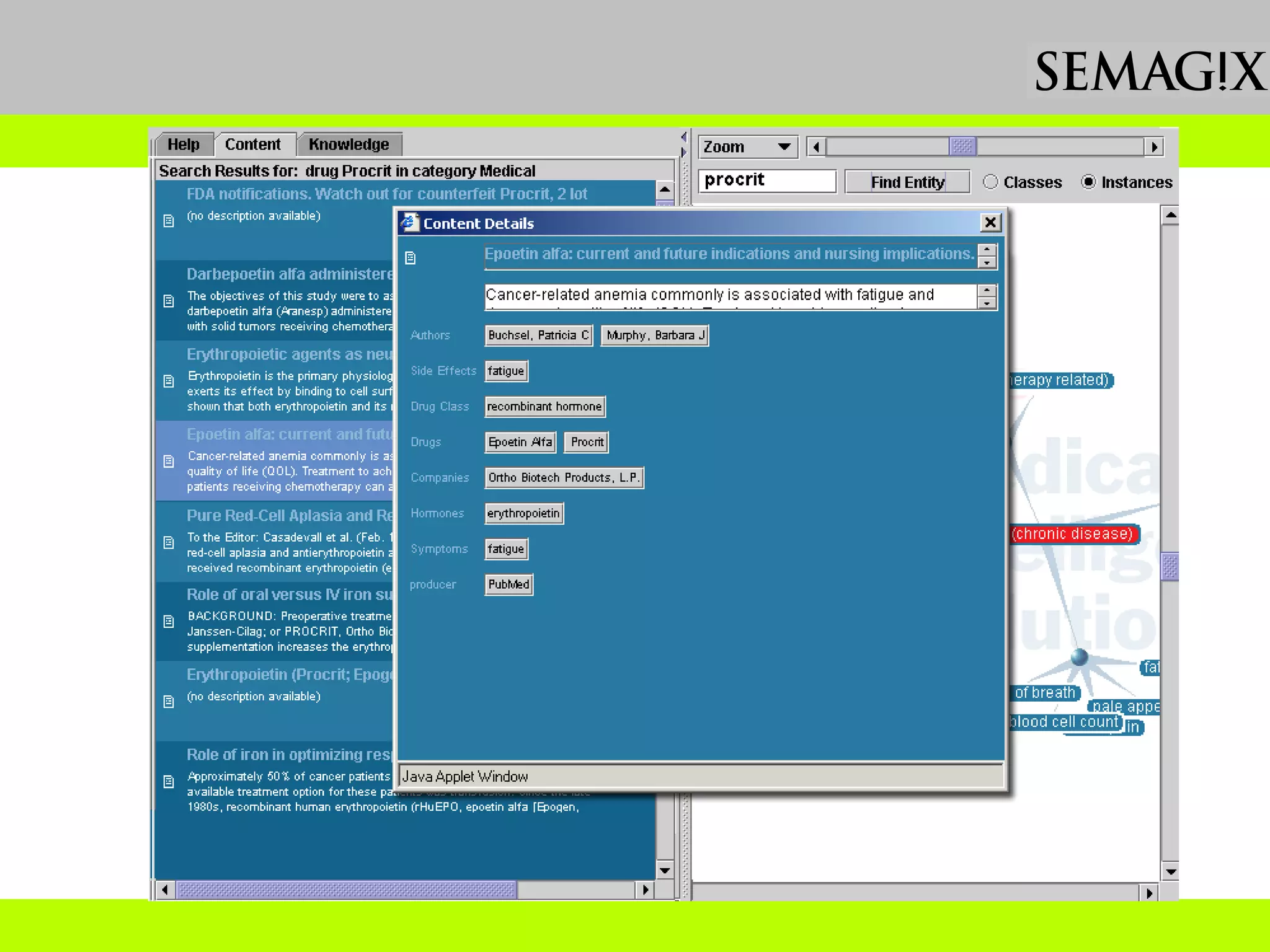
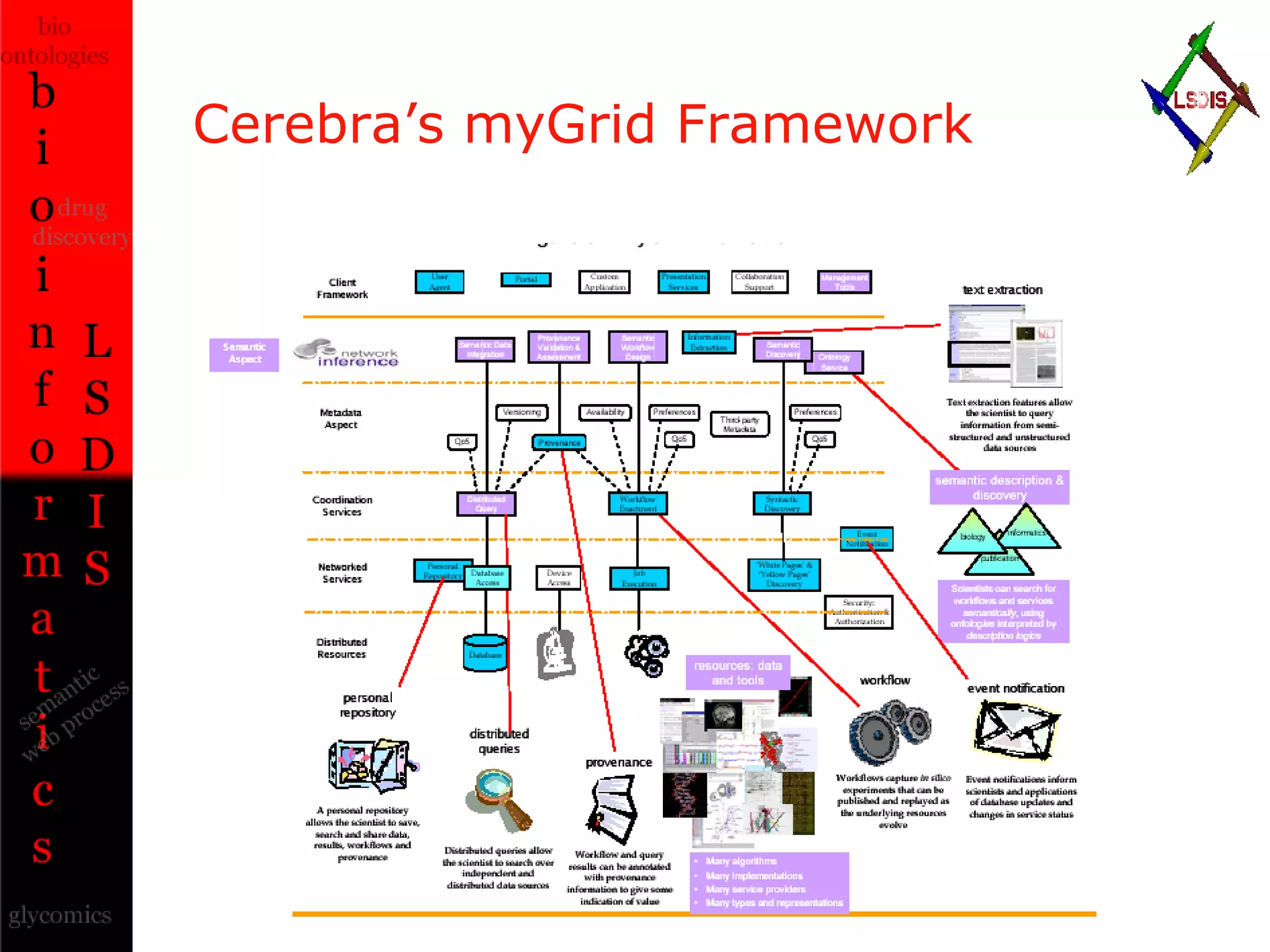
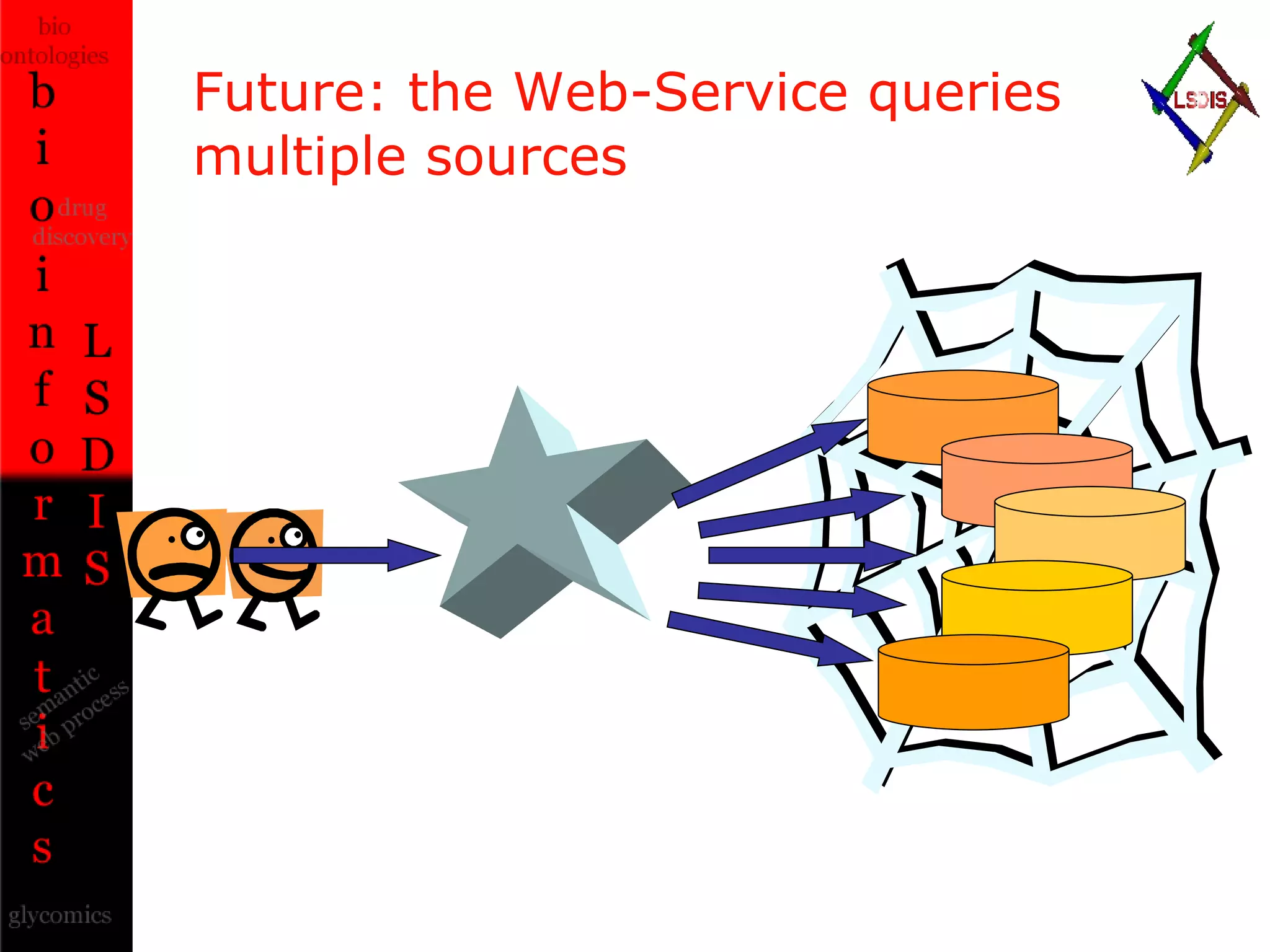
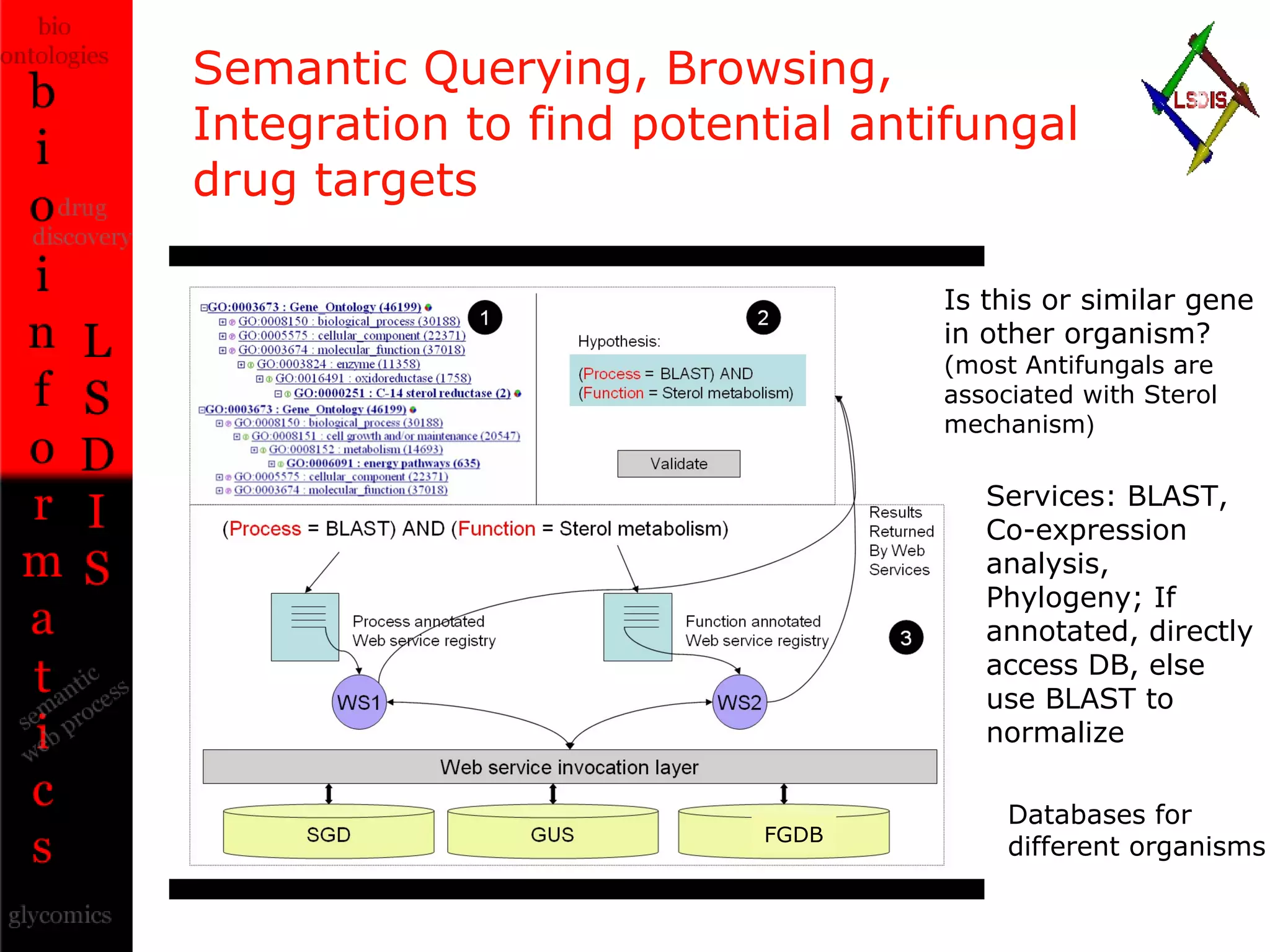
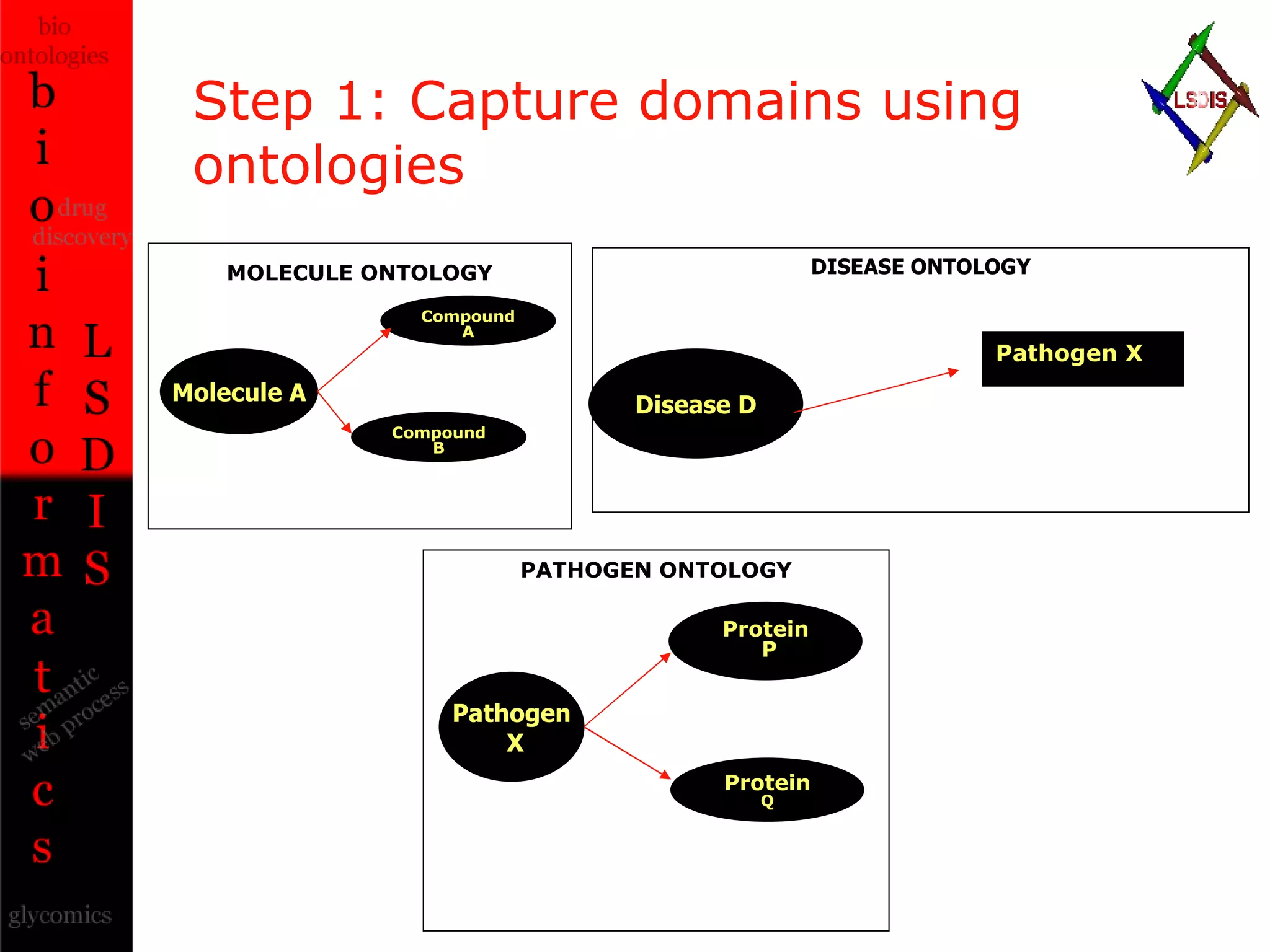
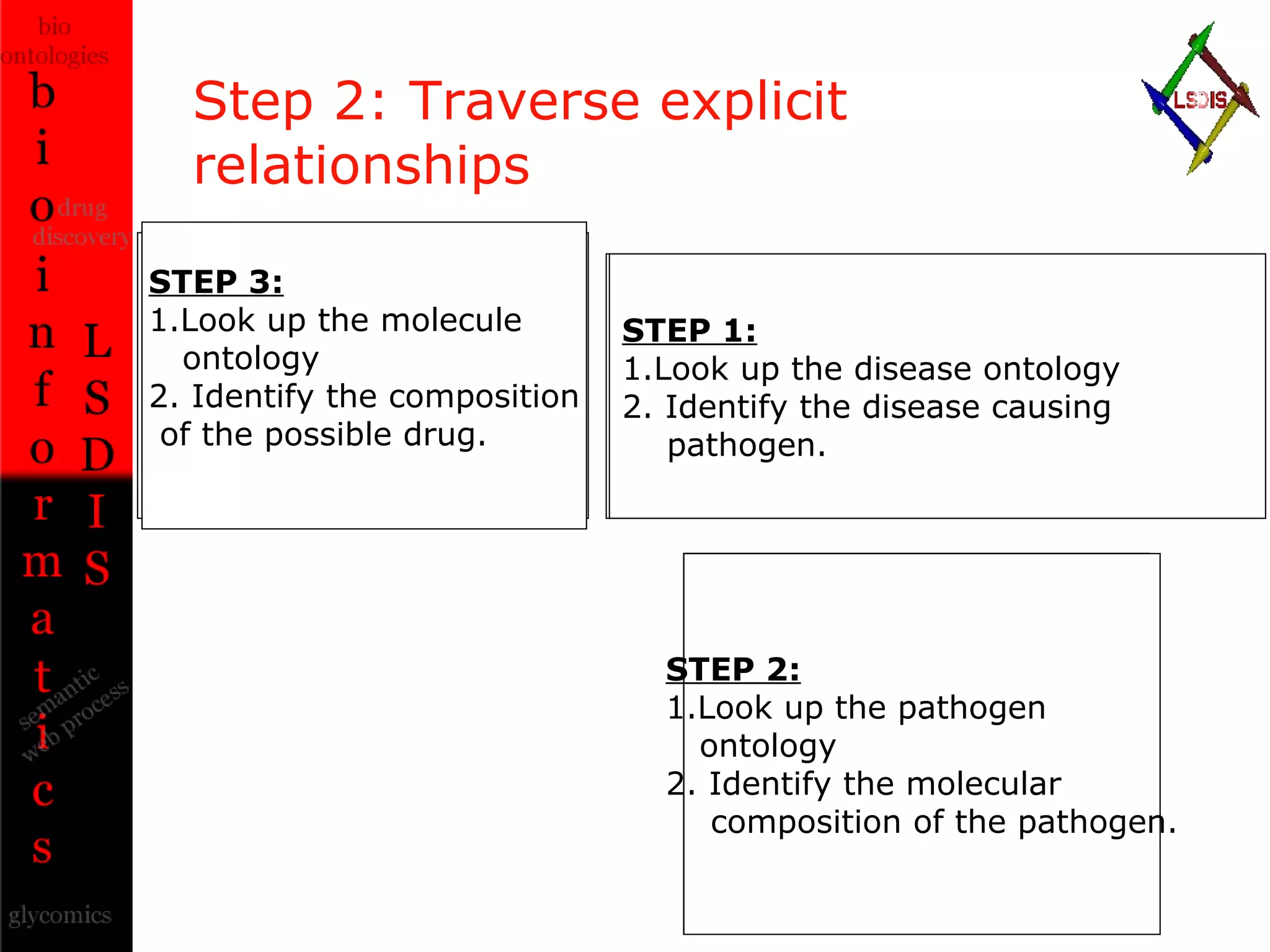
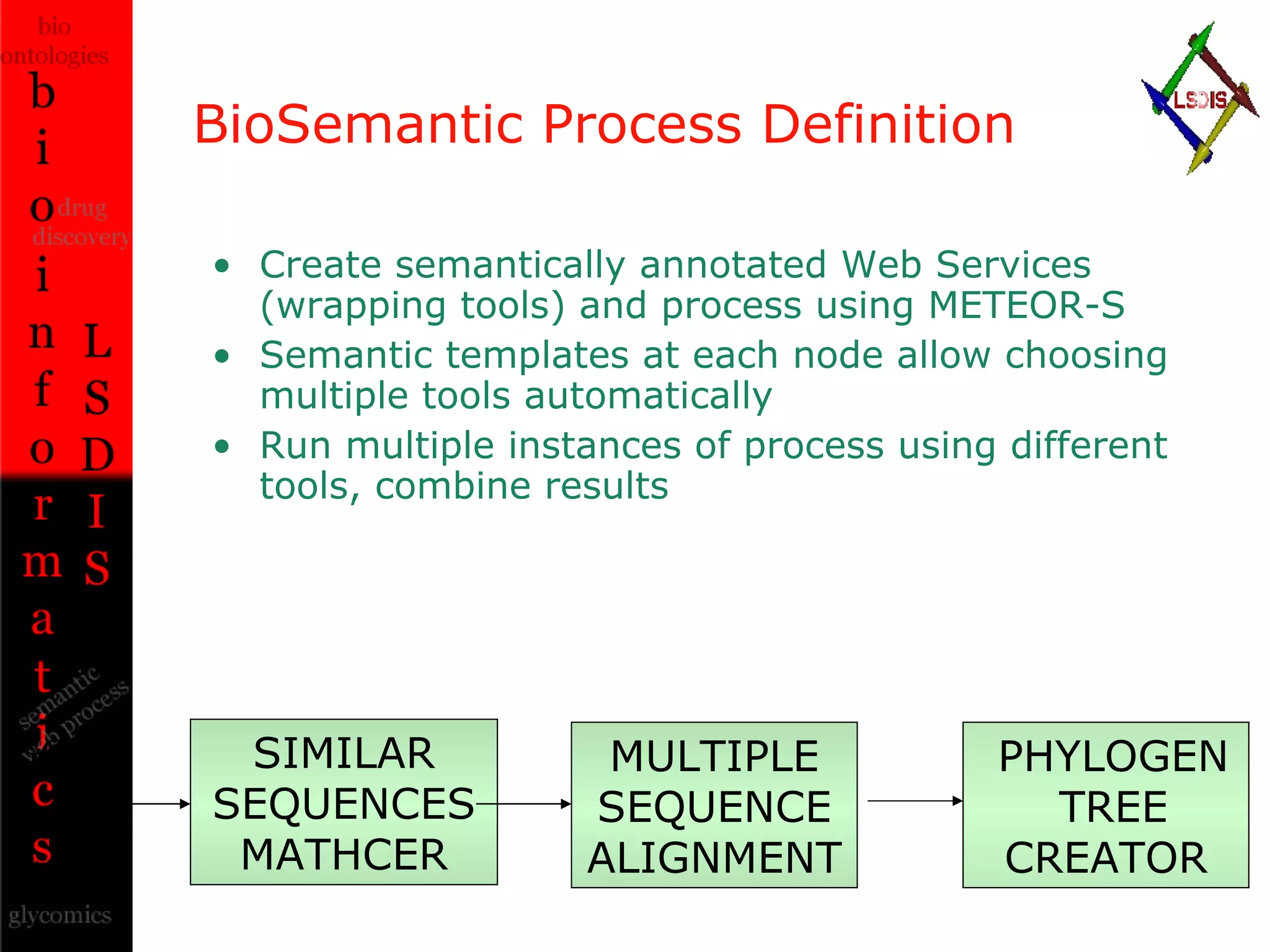
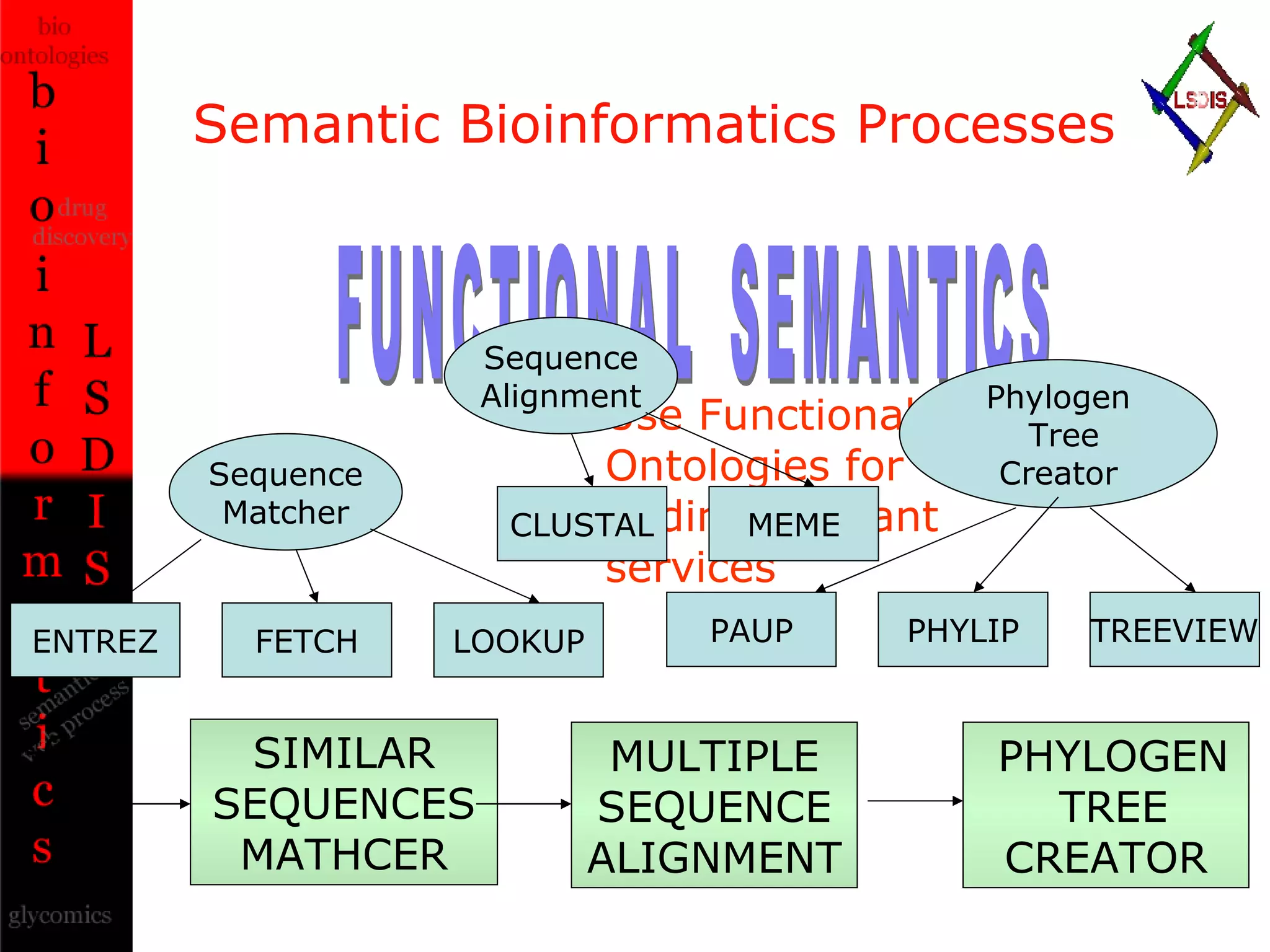
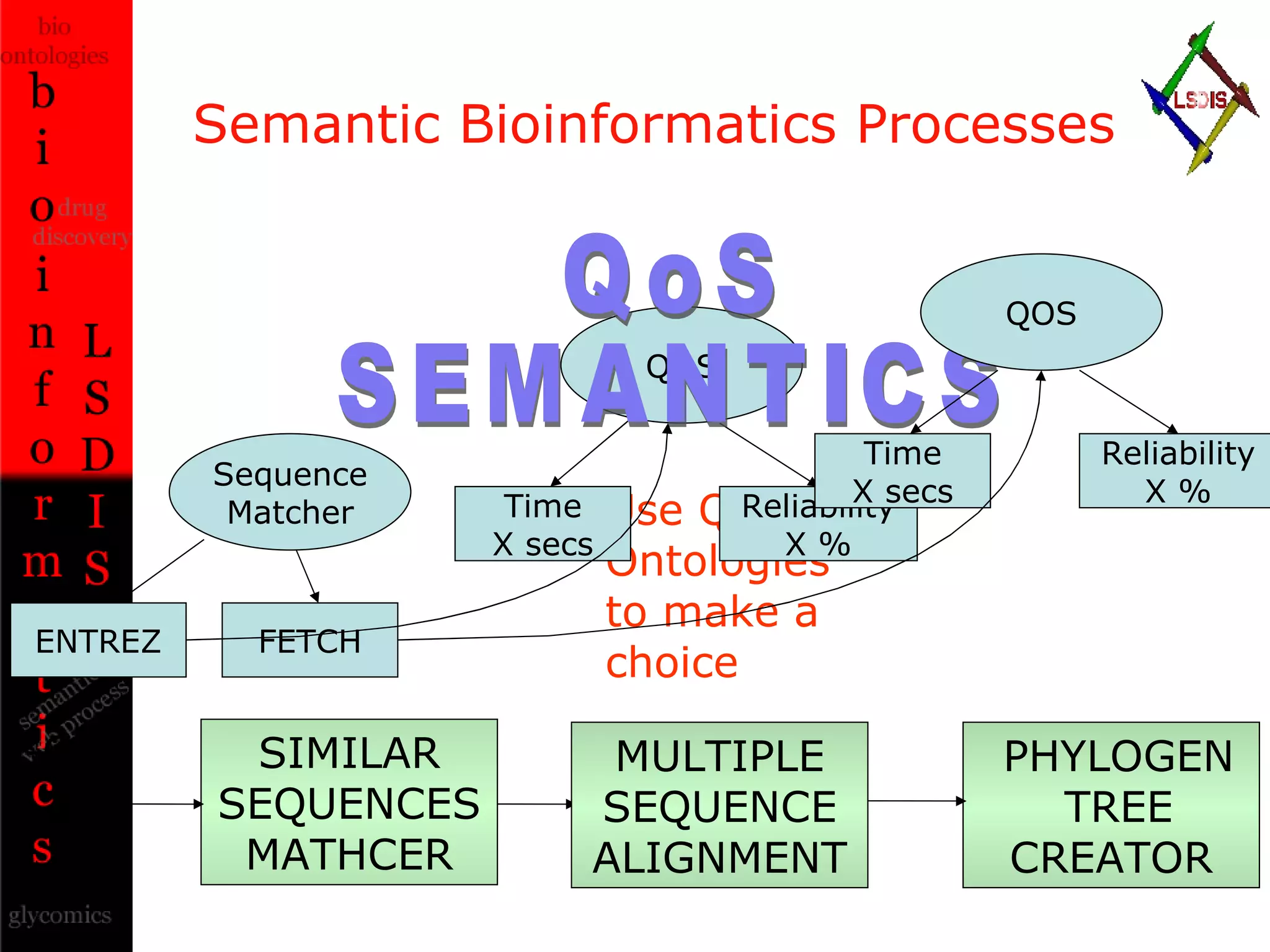
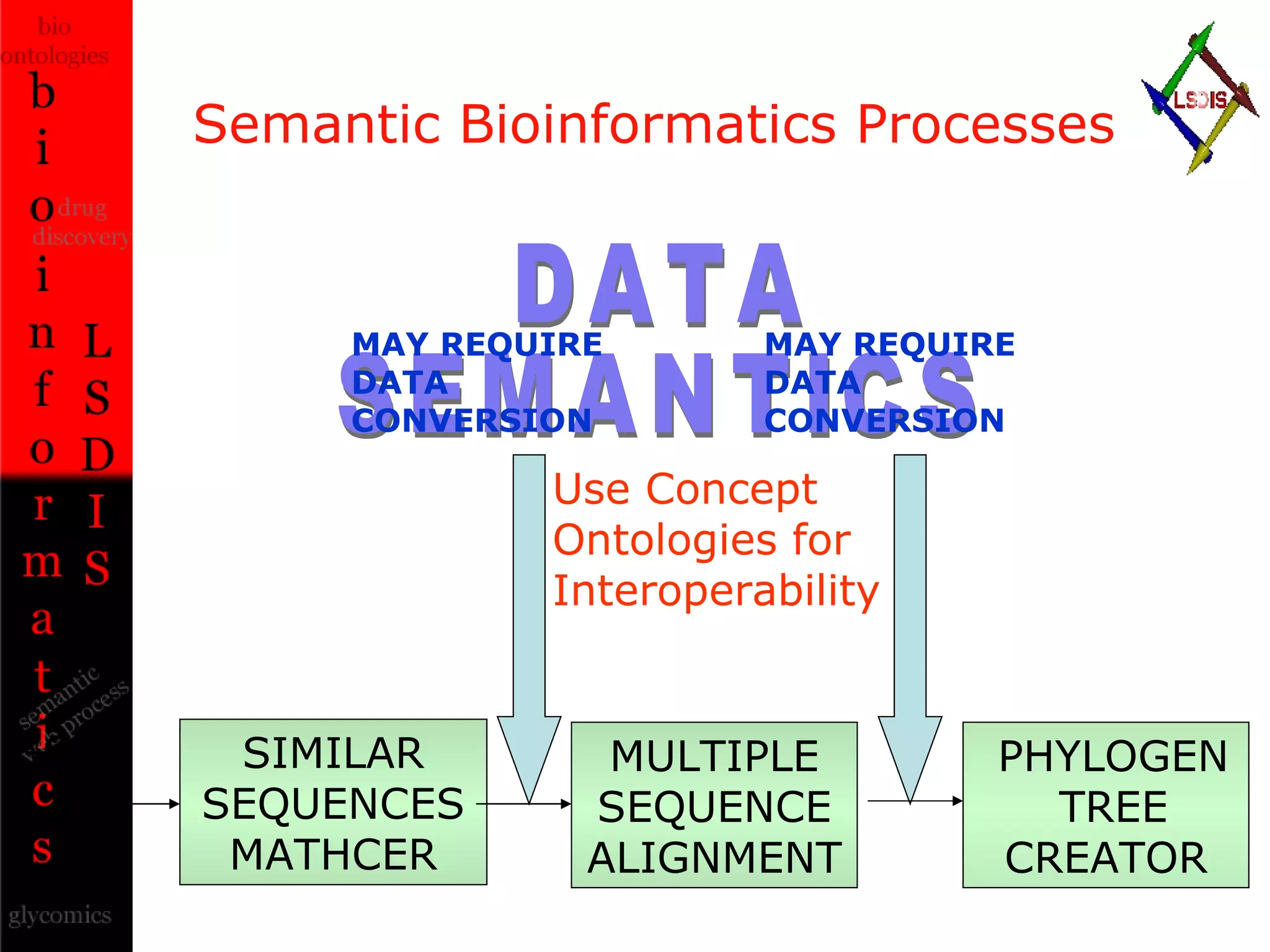
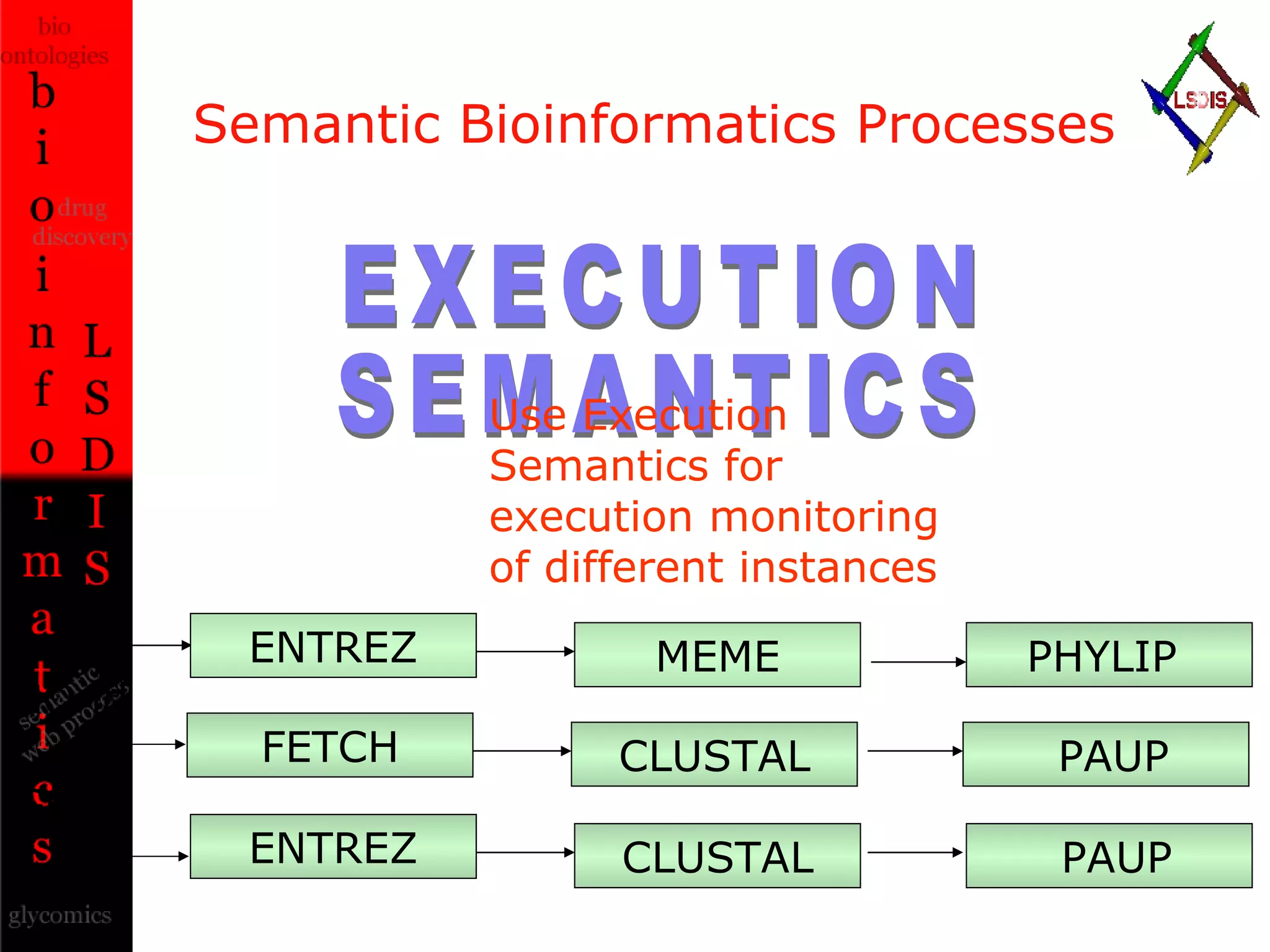
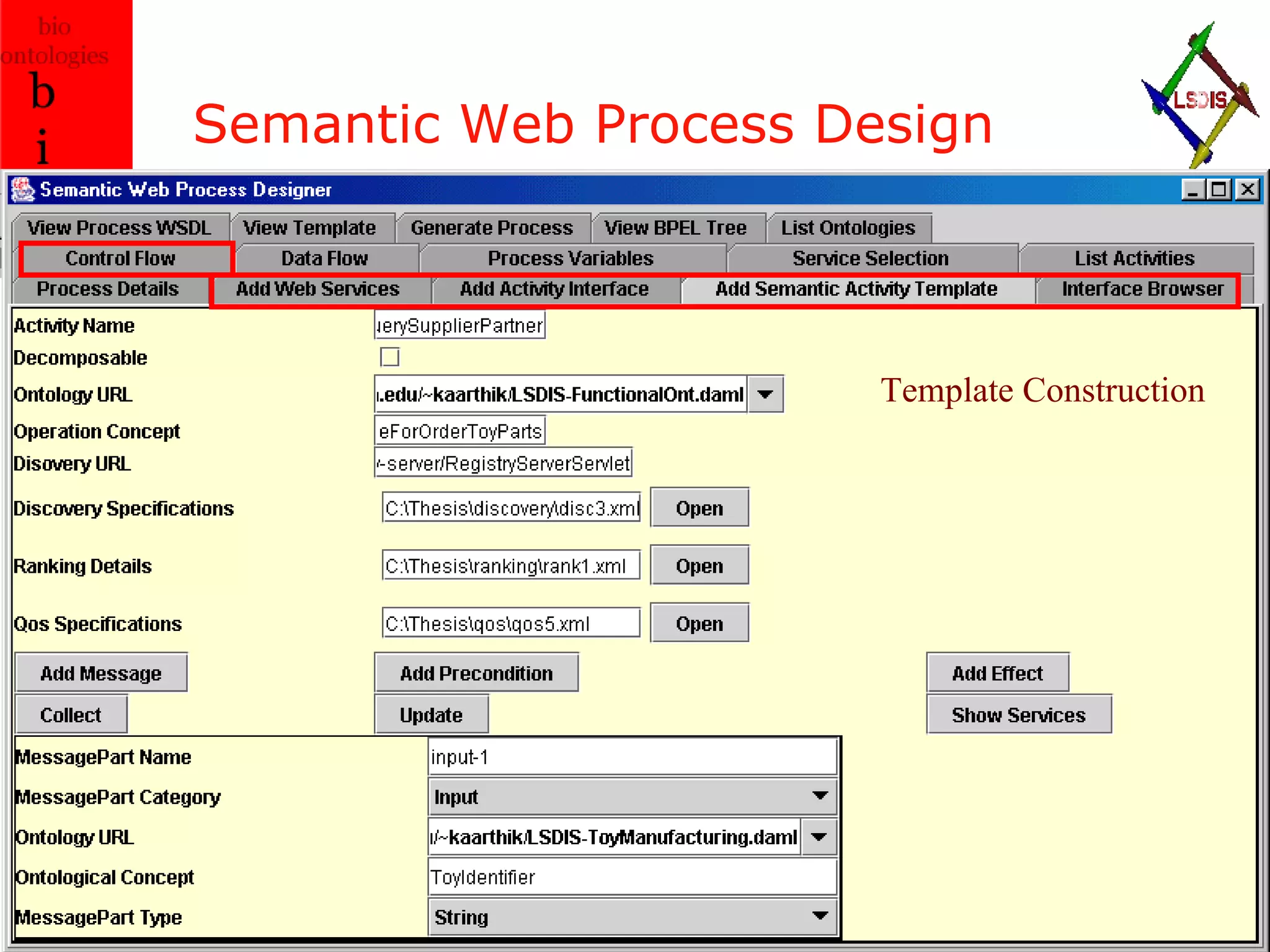
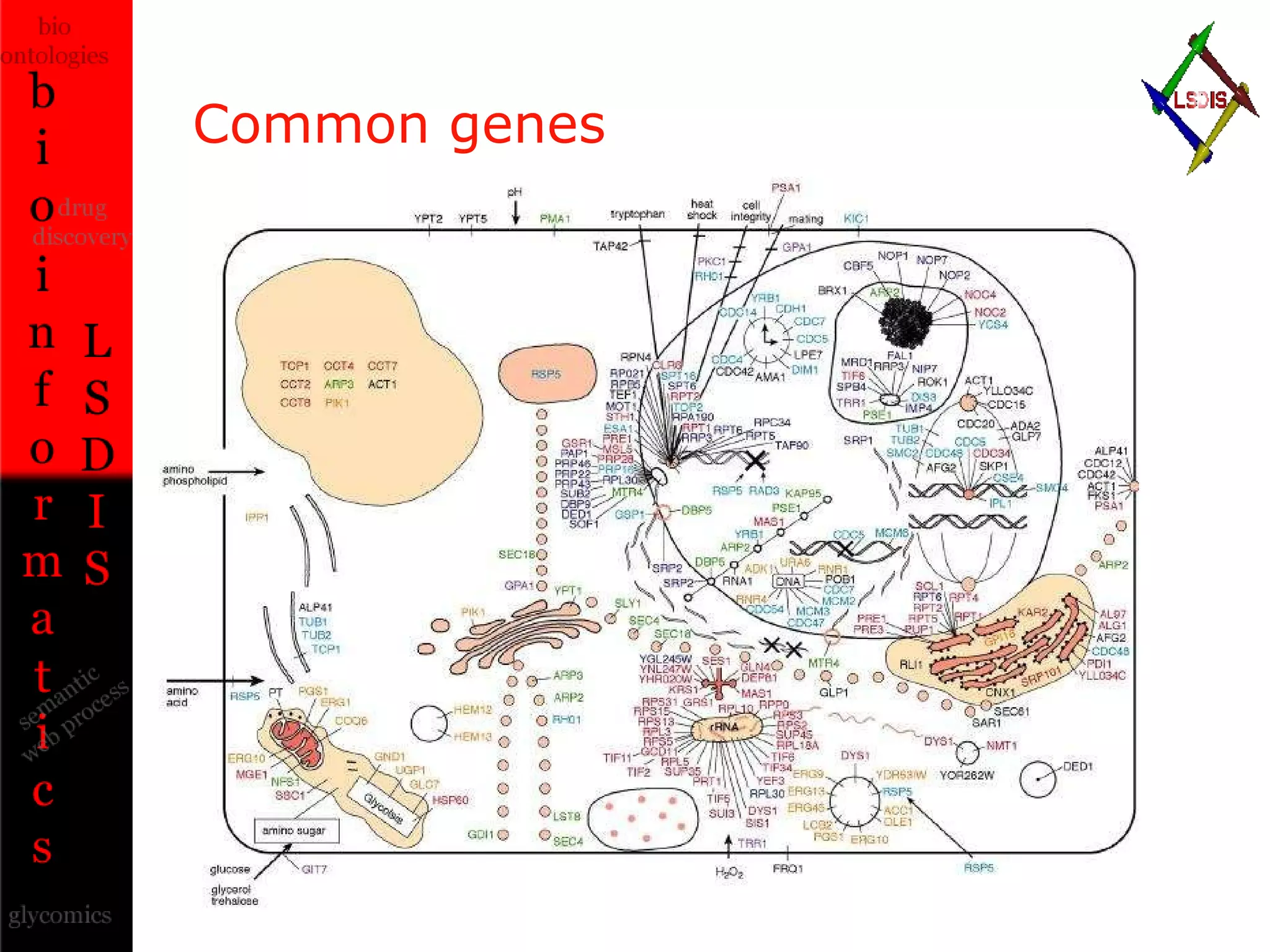
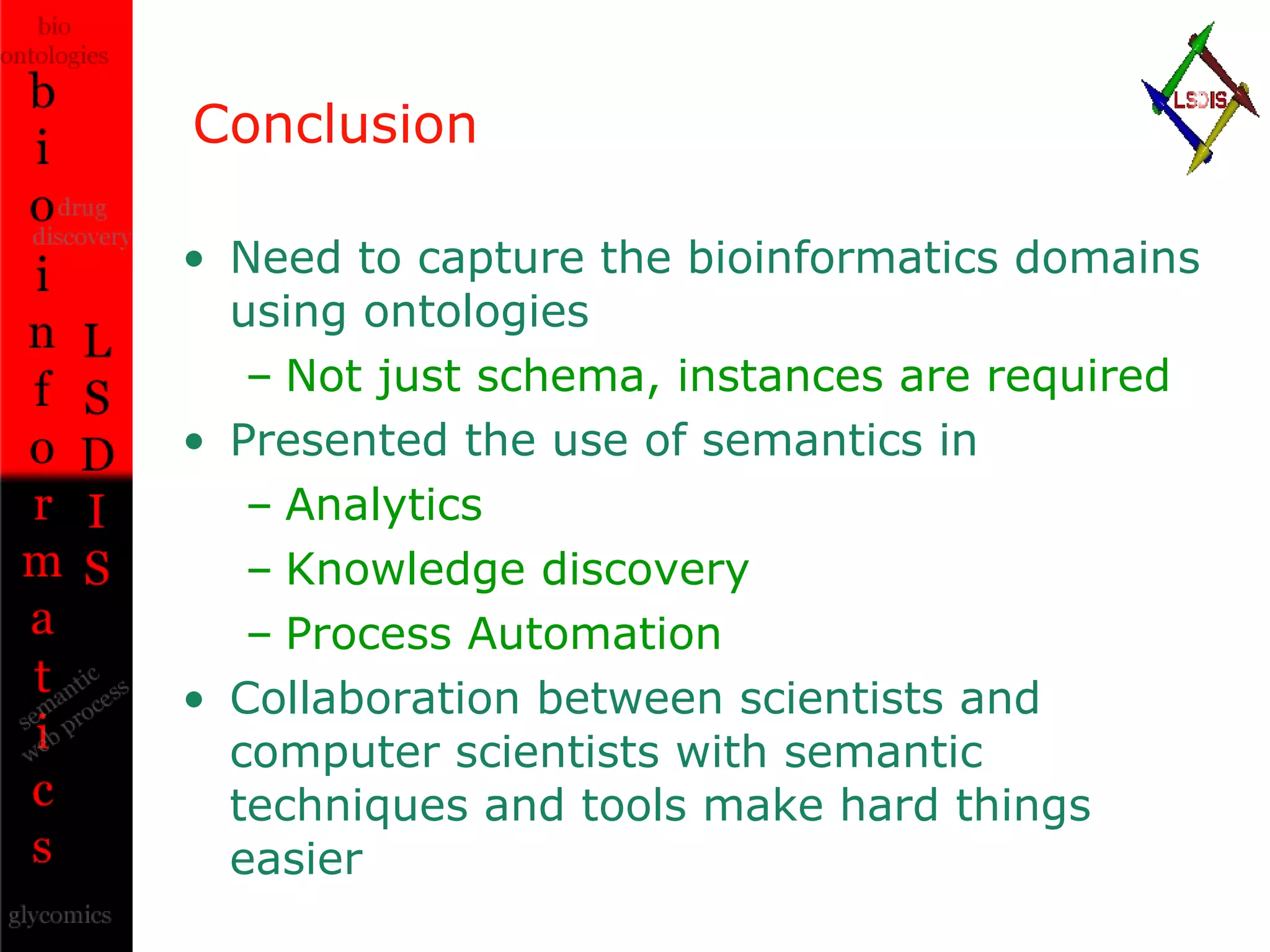
The document discusses the evolution and impact of bioinformatics, highlighting the role of semantics and ontologies in improving data integration and analysis in biological research. It emphasizes how semantic technologies can streamline processes, enhance knowledge discovery, and facilitate collaboration among scientists in various biological sub-fields. Additionally, it presents examples of applying semantic techniques in drug discovery and cancer research to expedite research outcomes.