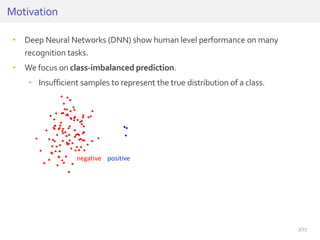
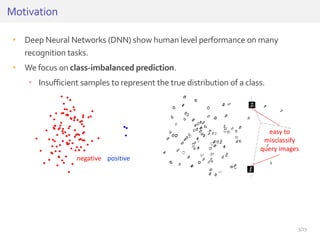
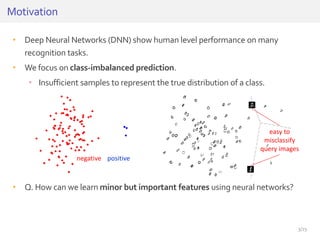
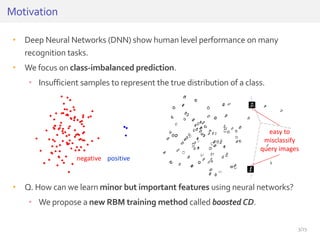
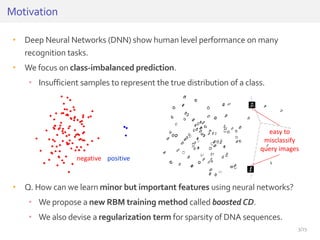
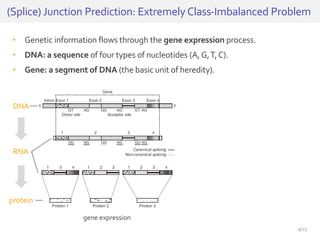
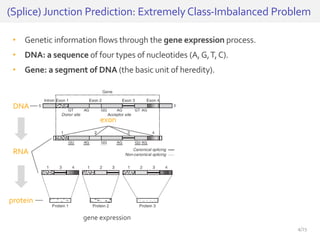
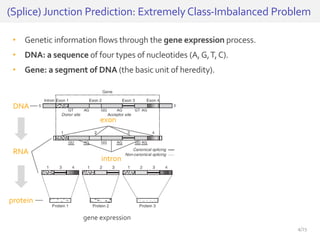
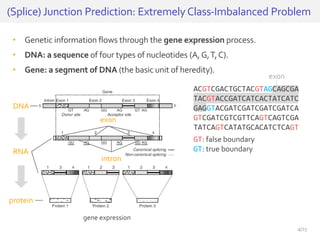
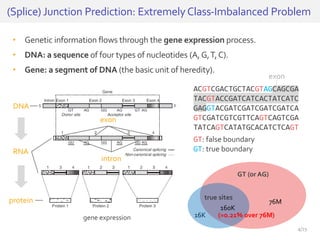
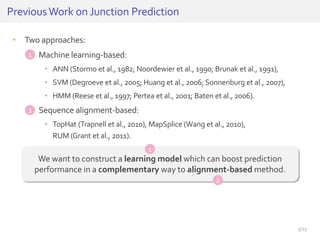
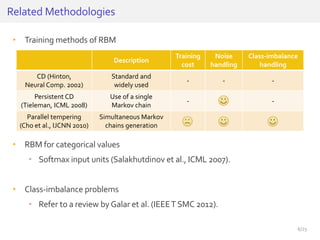
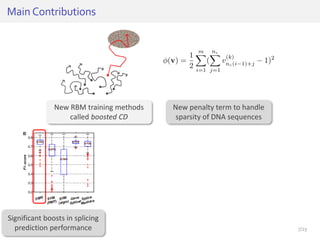
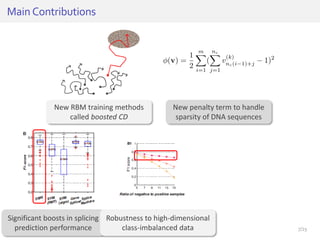
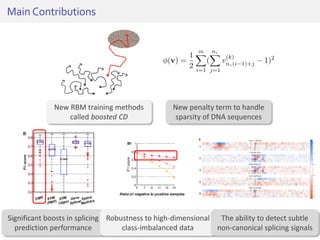
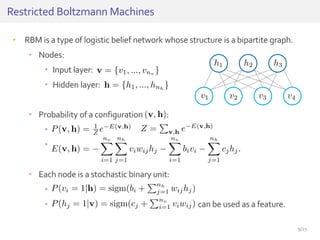
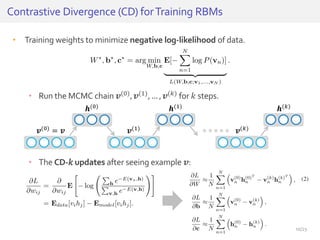
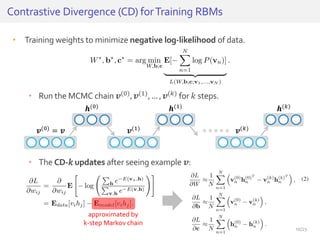
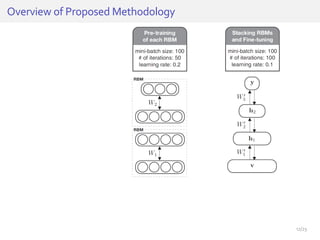
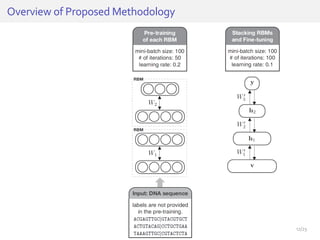
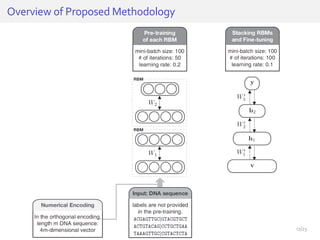
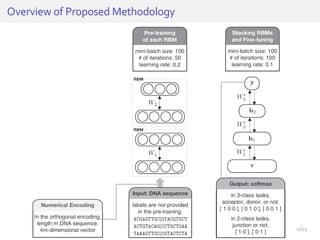
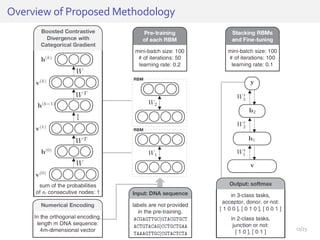
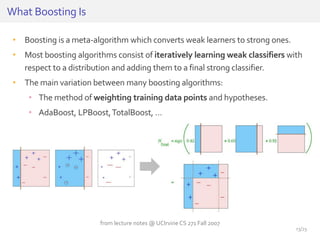
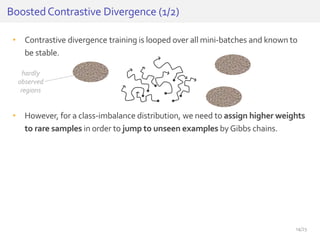
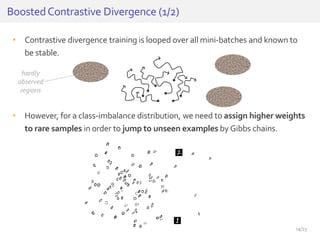
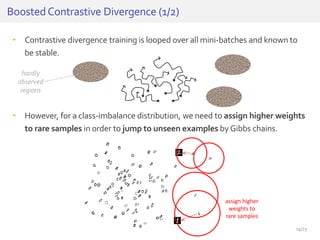
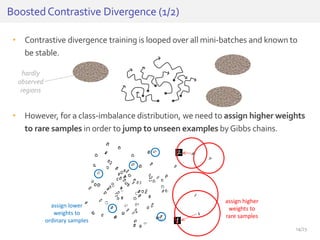
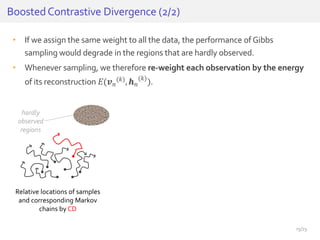
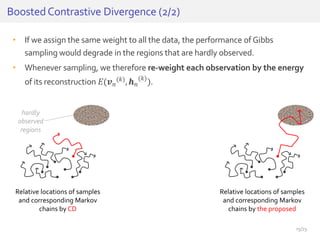
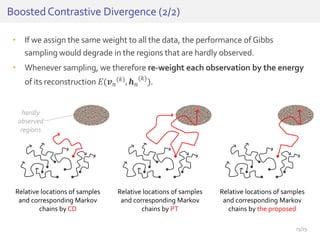
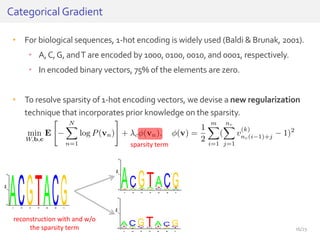
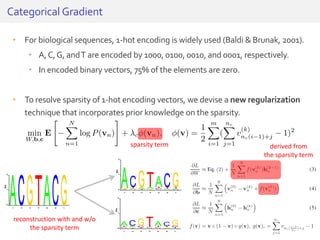
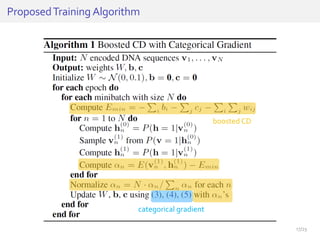
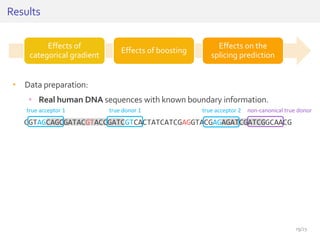
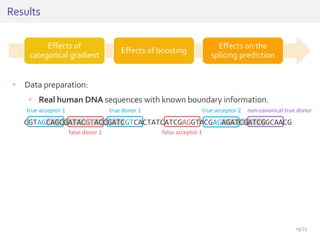
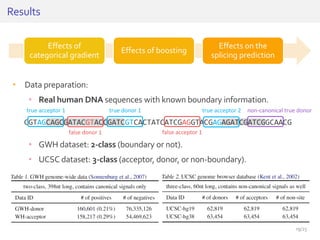
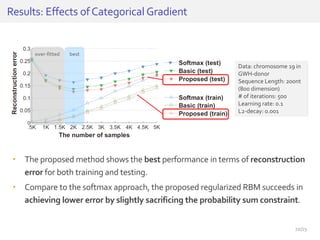
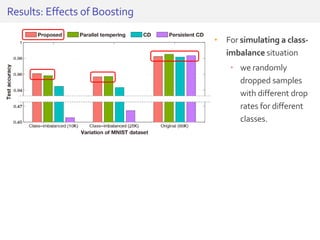
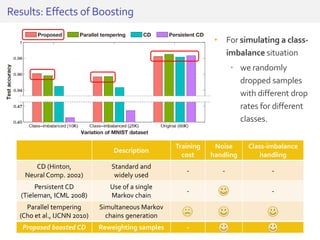
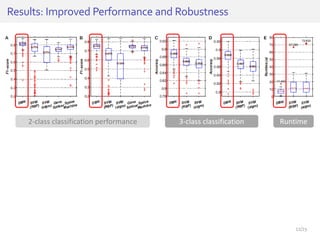
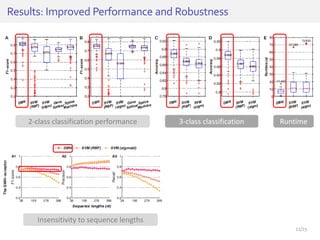
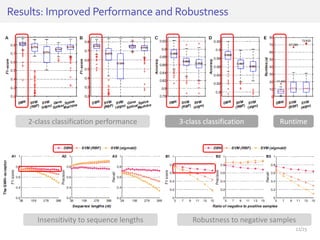
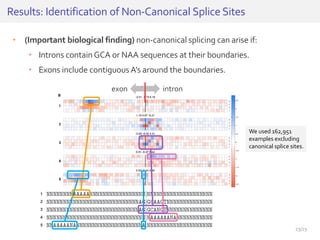
The document discusses a novel approach for splice junction prediction using a boosted categorical restricted Boltzmann machine to enhance performance in class-imbalanced datasets, particularly relevant for deep neural networks. It presents new training methods, including boosted contrastive divergence and a regularization term for sparsity in DNA sequences, showing significant improvements in prediction accuracy. Additionally, the study compares its methodology to existing machine learning and sequence alignment-based approaches, demonstrating robustness in handling high-dimensional data.