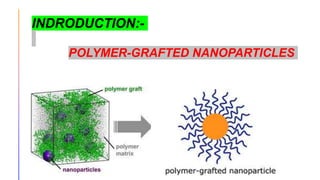
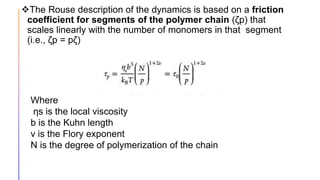
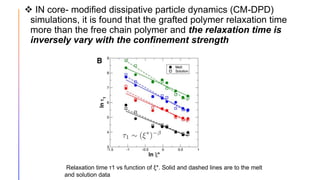
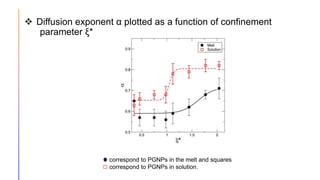
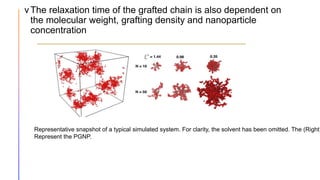
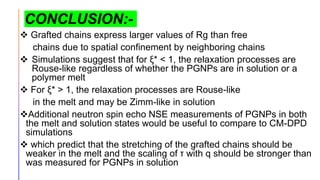
The document discusses the simulation of polymer-grafted nanoparticles (PGNPs) to study the dynamics of polymer chains. The simulation finds that the relaxation time of polymer chains near the nanoparticle surface is longer than those further away due to spatial confinement. It also finds that the relaxation time of grafted polymers is longer than free polymers and decreases with increased confinement. The relaxation processes of PGNPs are found to be Rouse-like in polymer melts and may be Zimm-like in solutions, depending on the confinement parameter. Additional neutron spin echo measurements are suggested to further validate the simulations.