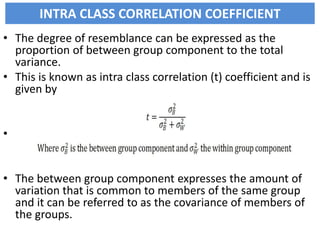
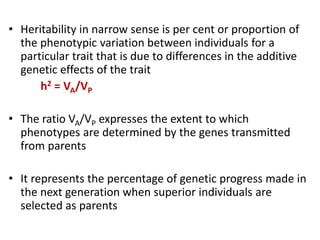
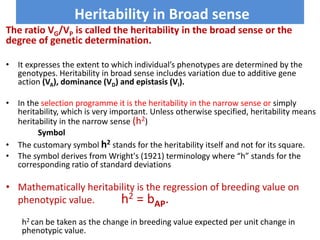
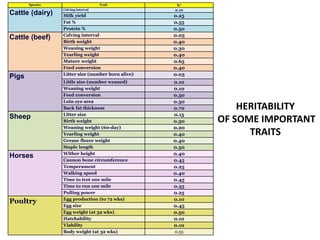
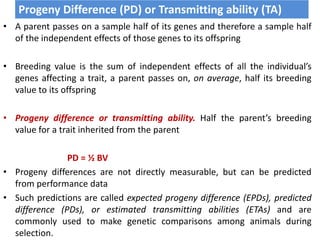
This document discusses resemblance between relatives and heritability. It states that relatives resemble each other due to common genes, and the degree of resemblance can be used to estimate additive genetic variance, which is heritable. Heritability is defined as the ratio of additive genetic variance to phenotypic variance, and represents the proportion of phenotypic variation attributable to genetics. High heritability traits are more responsive to selection. The document provides examples of heritability estimates for important traits in cattle, pigs, sheep, and poultry.