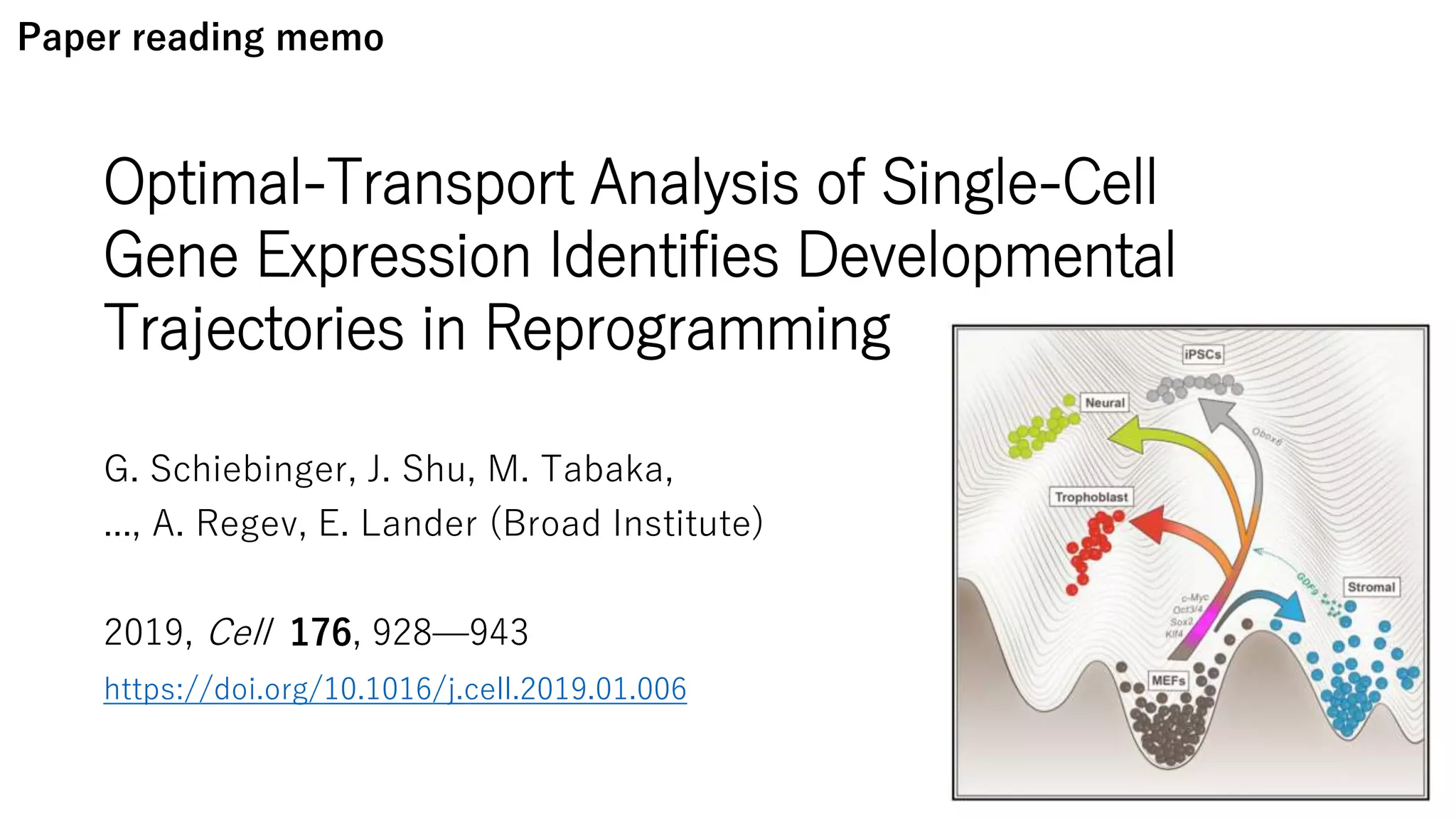
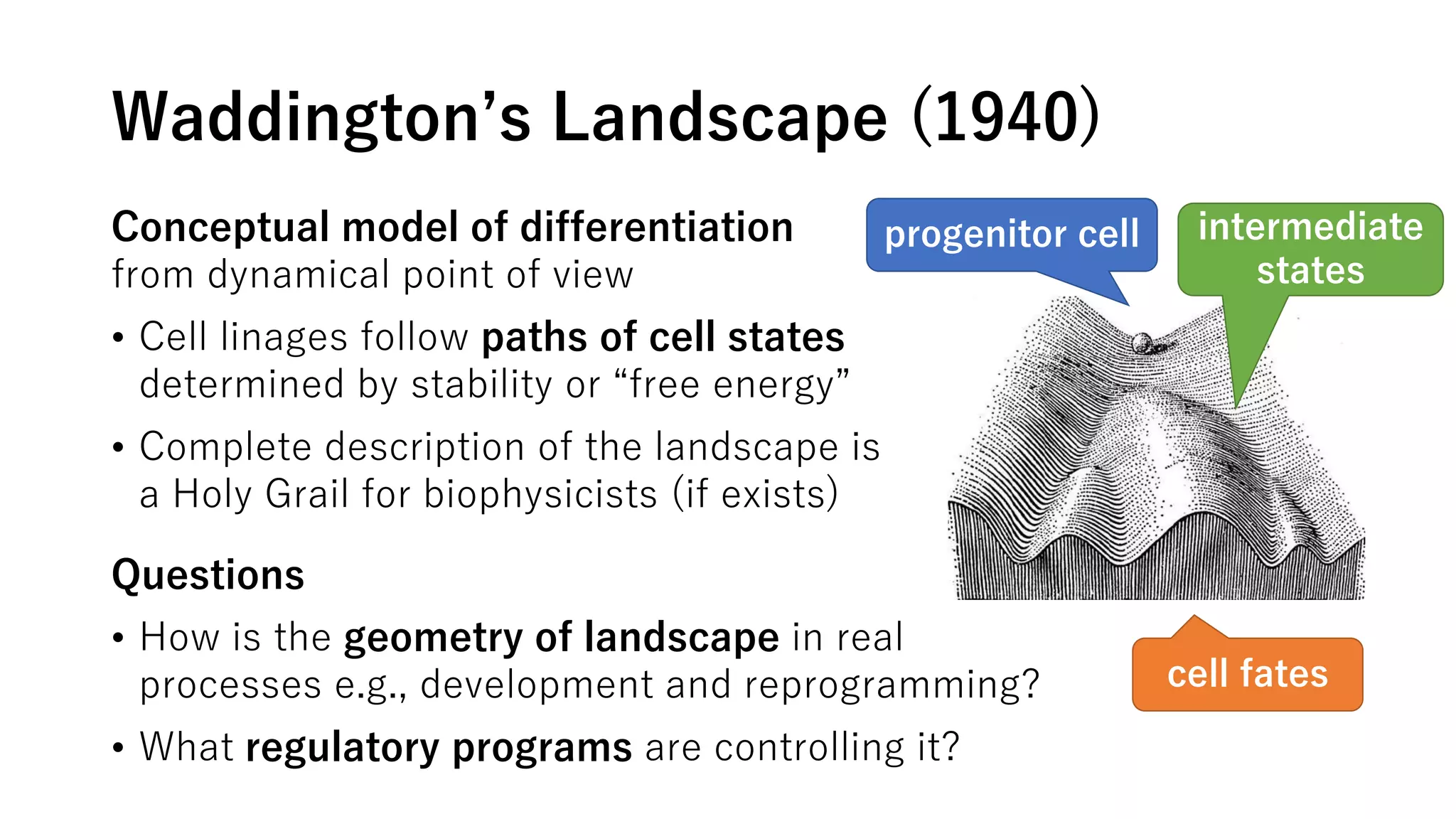
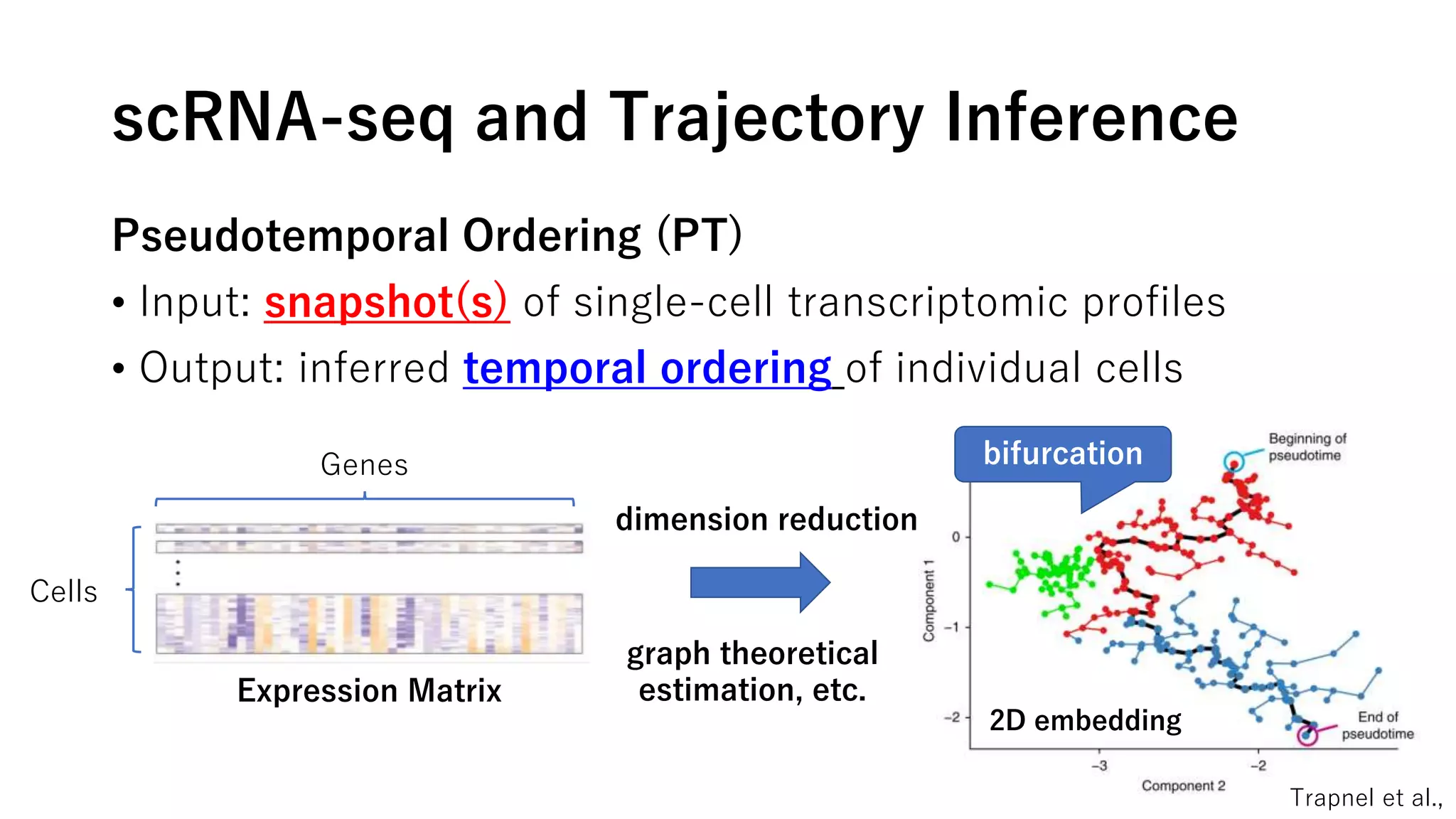
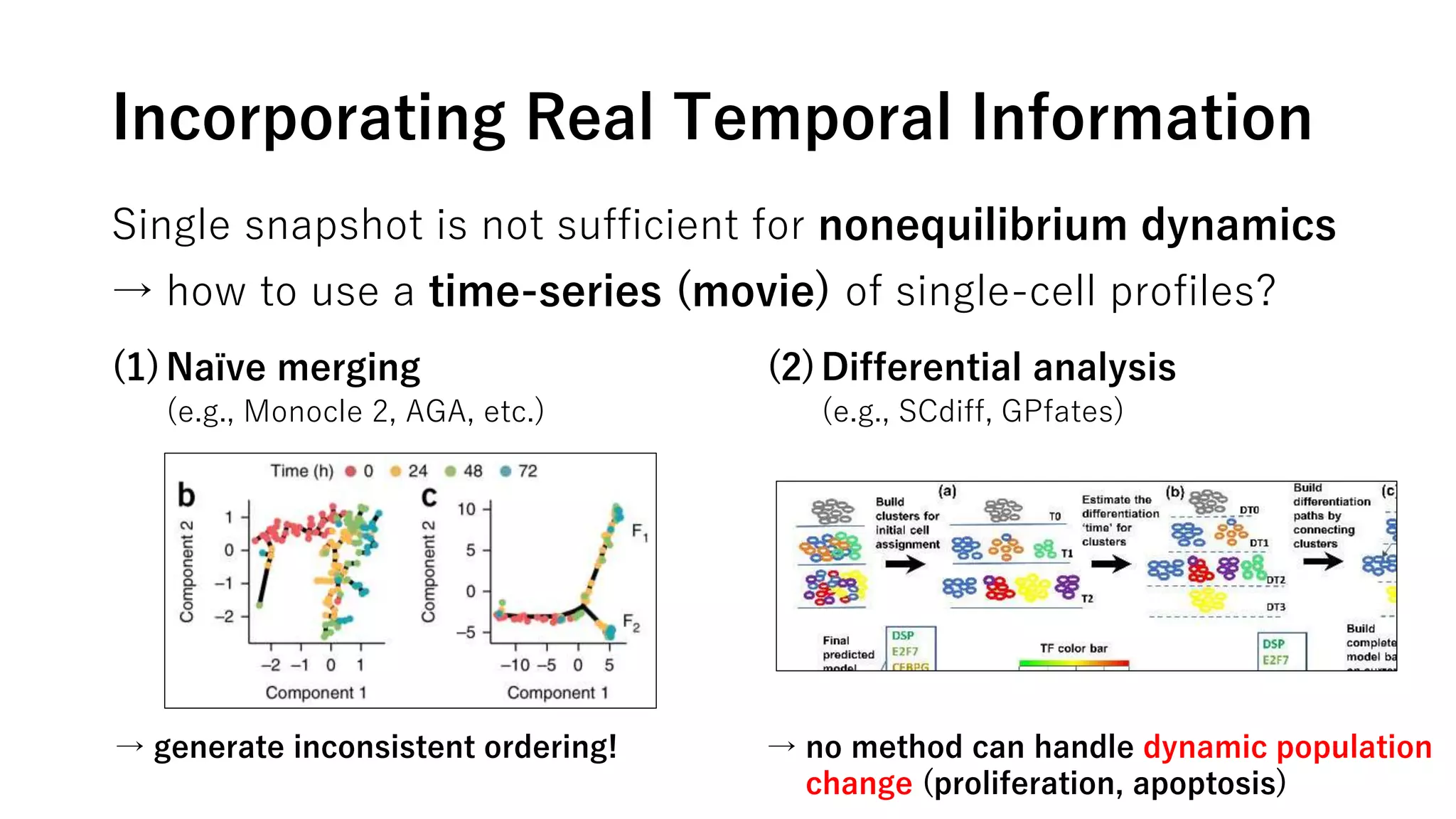
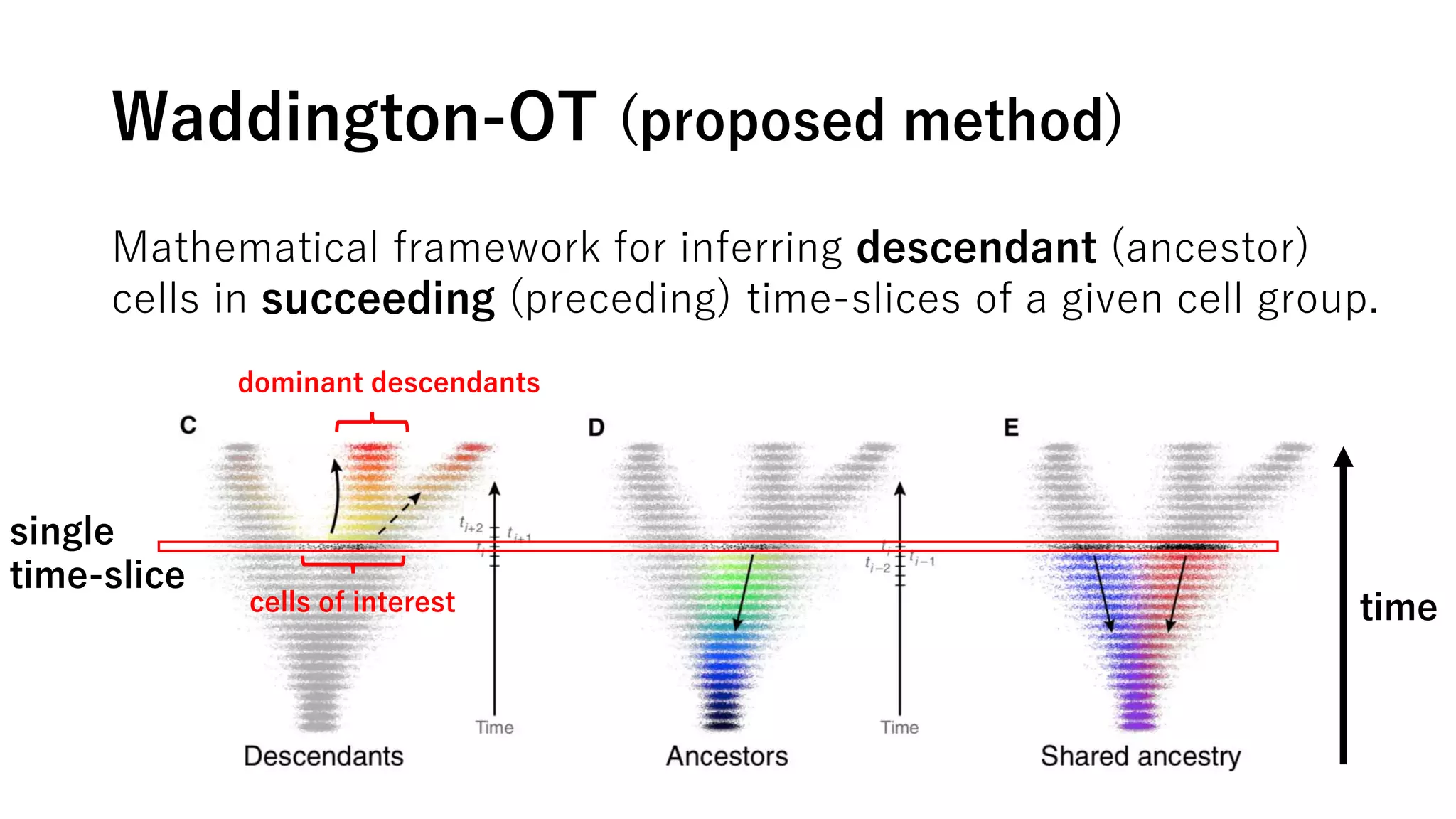
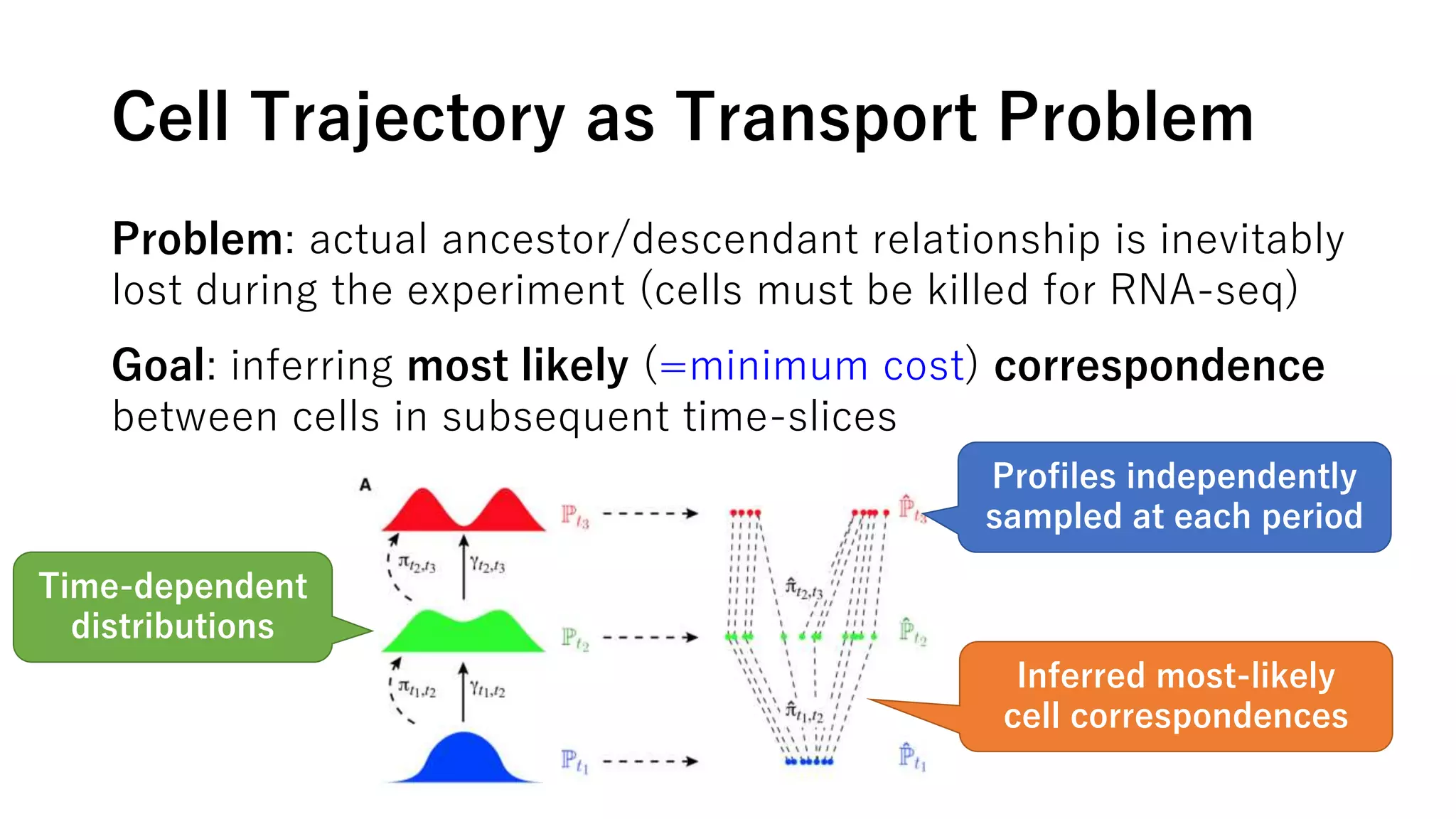
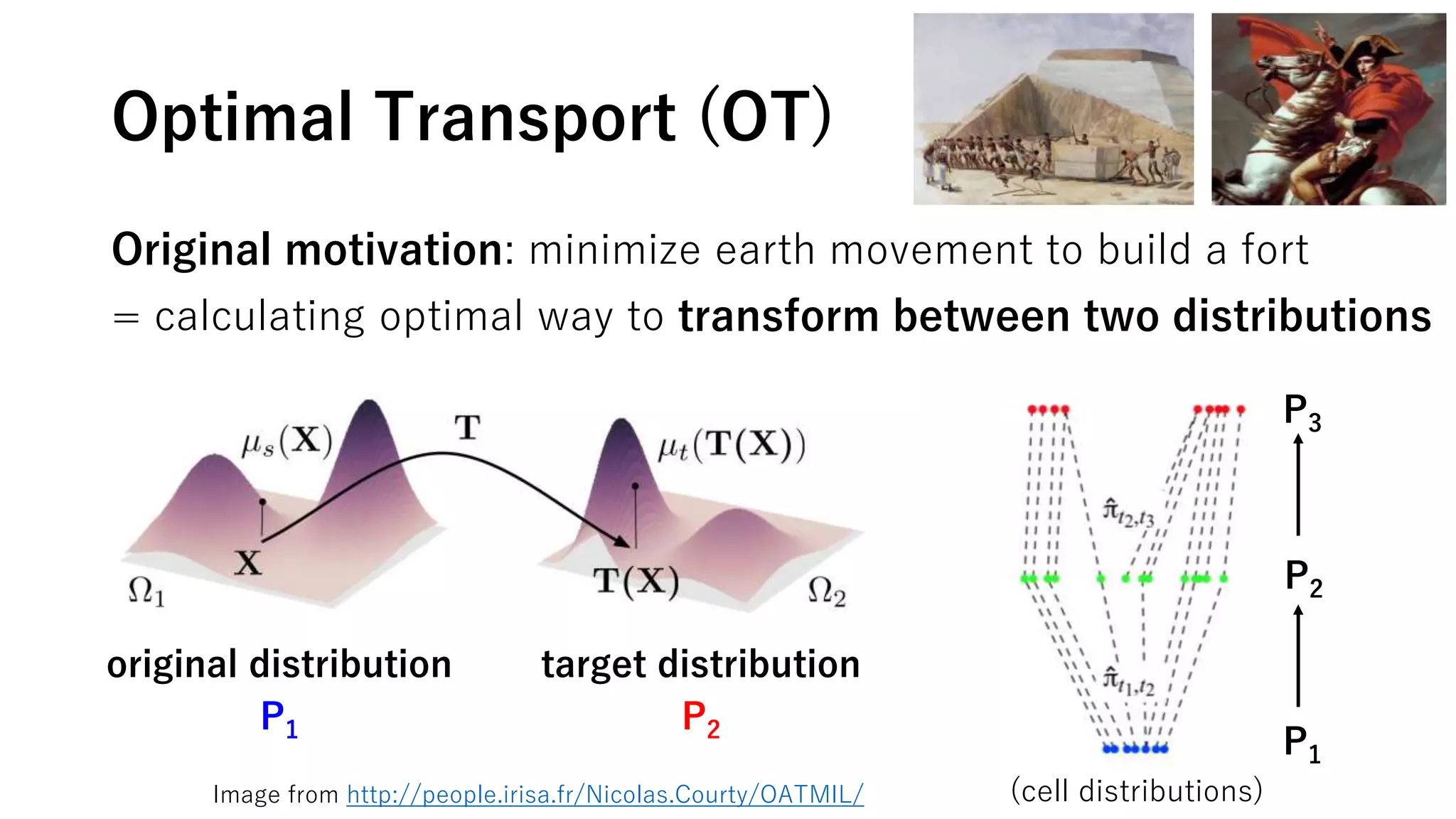
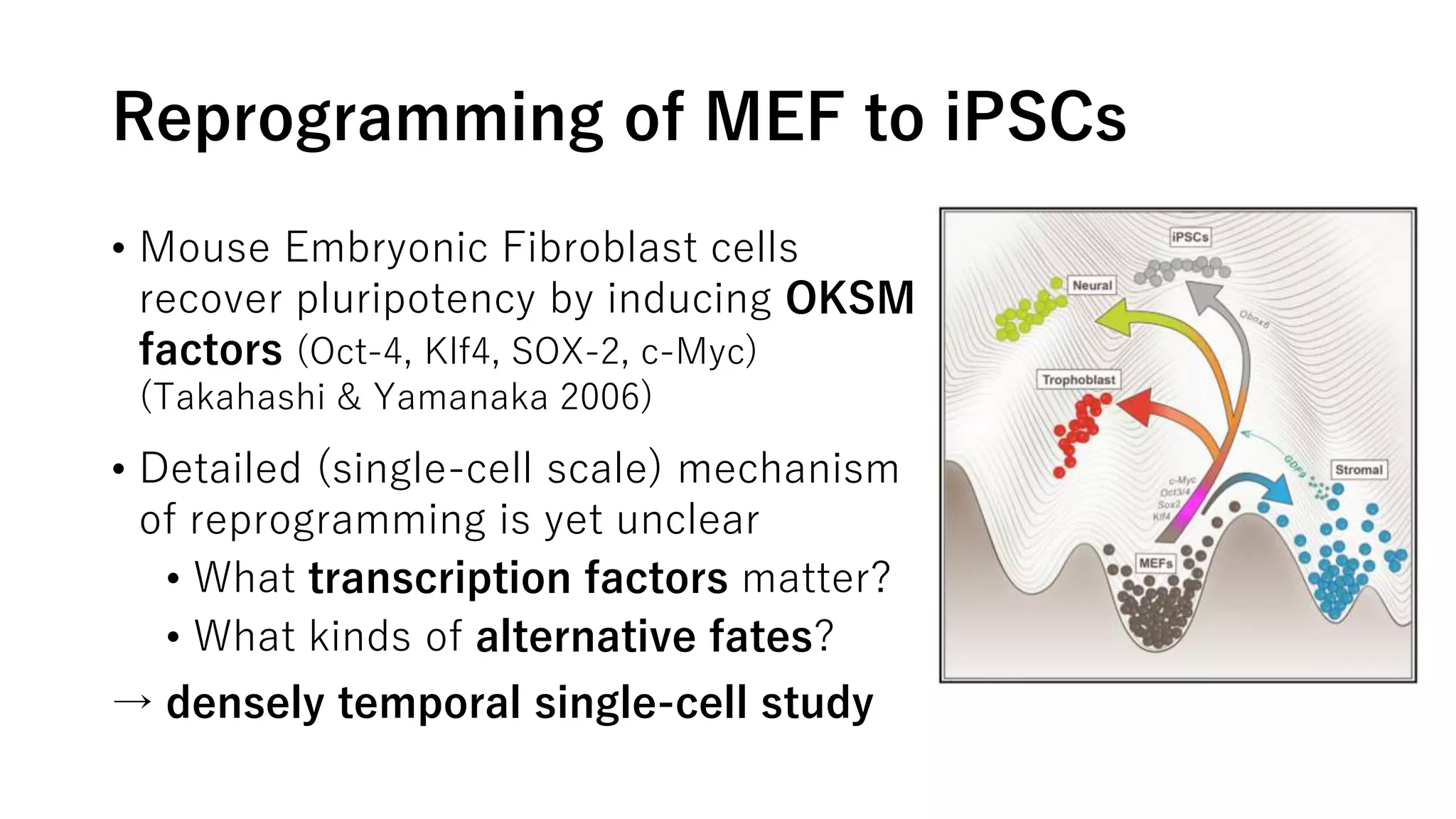
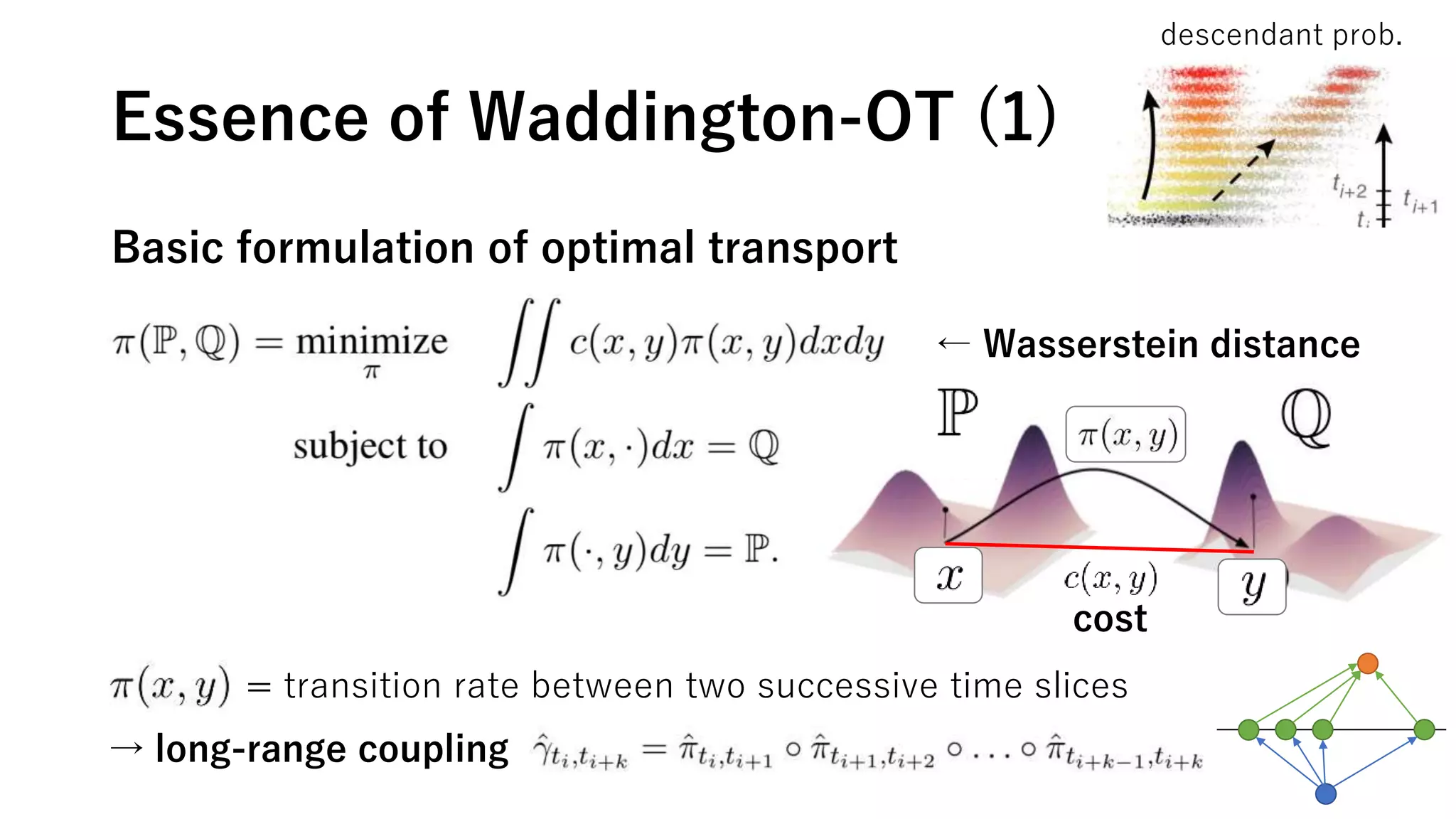
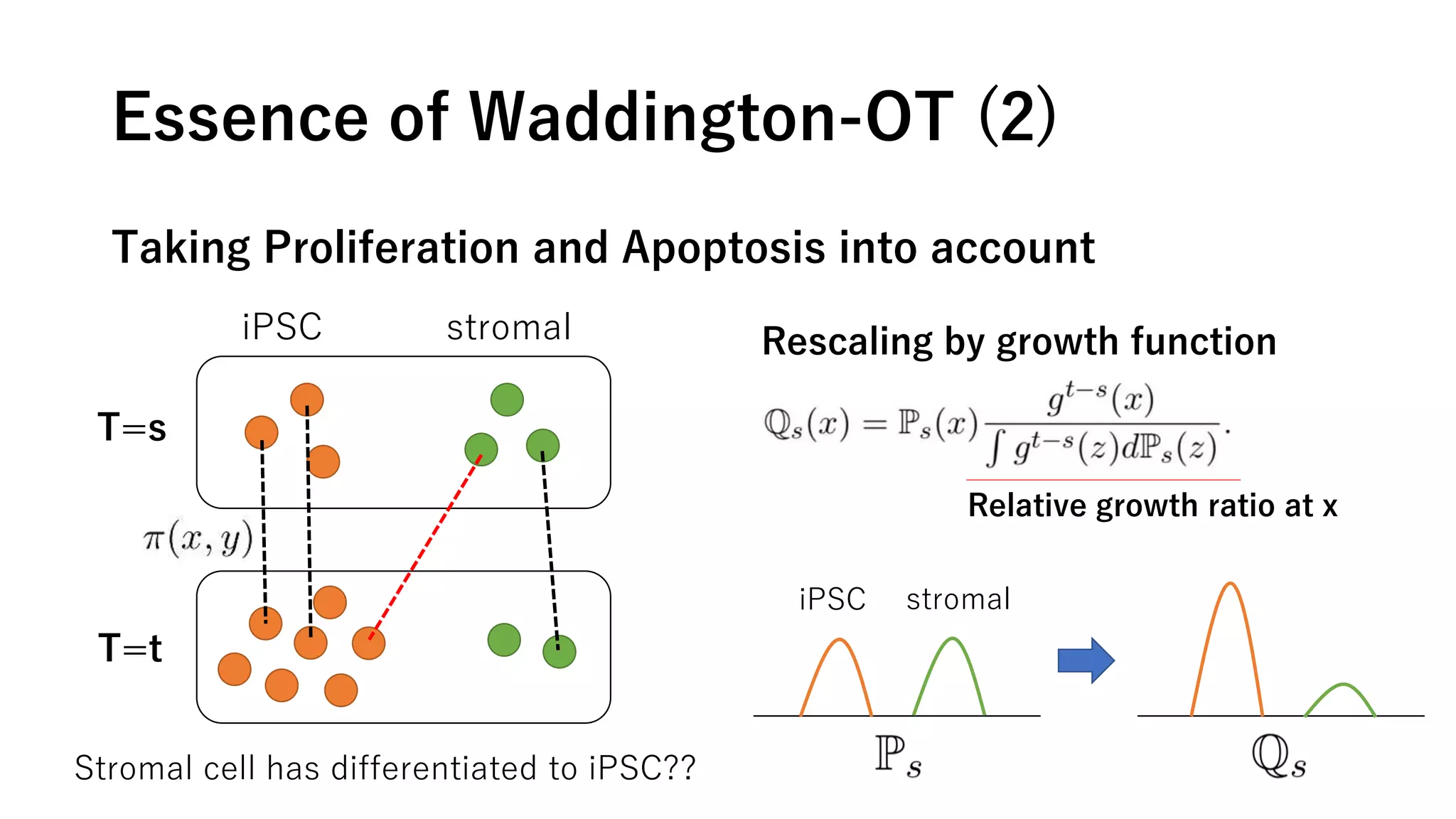
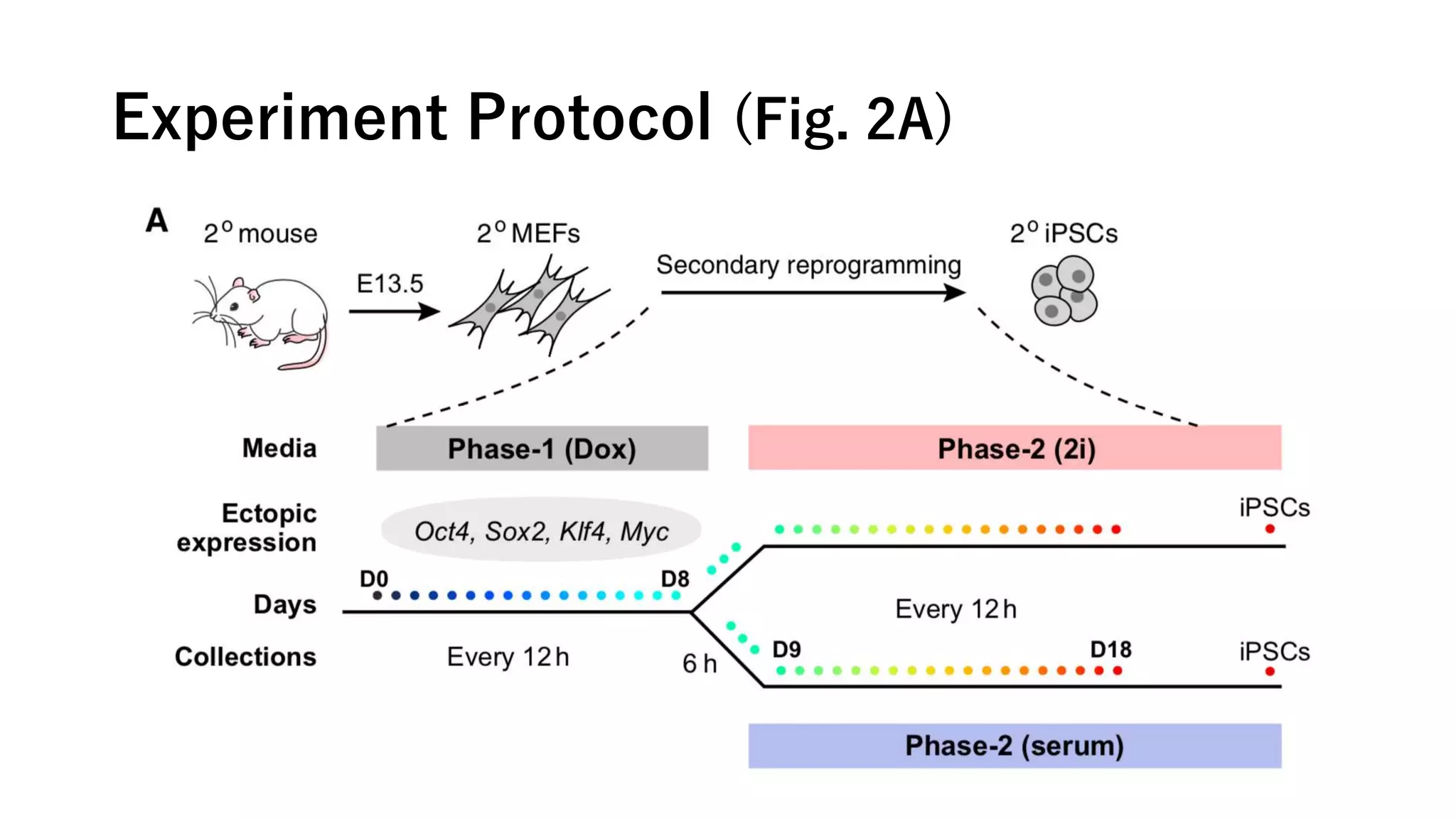
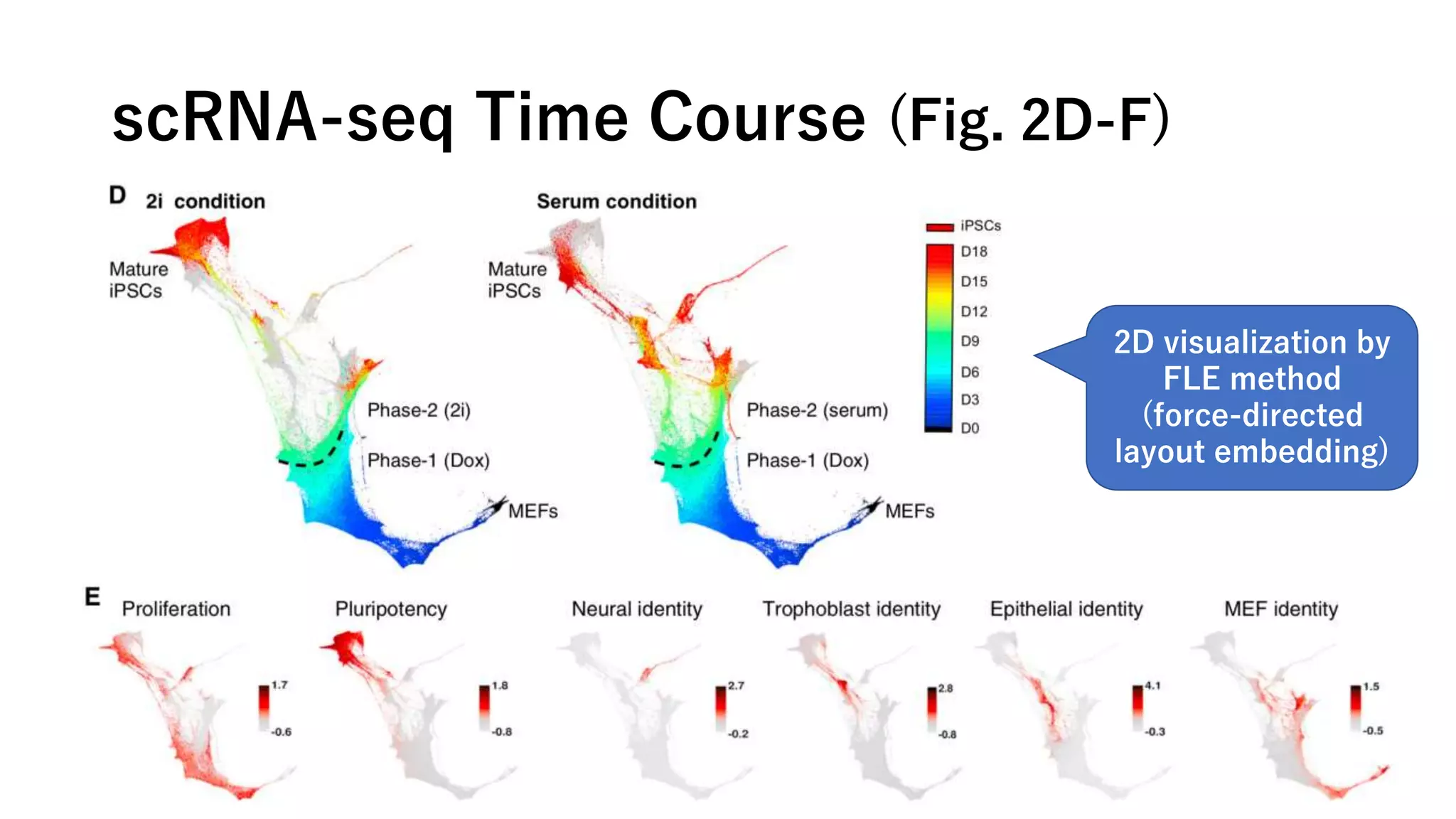
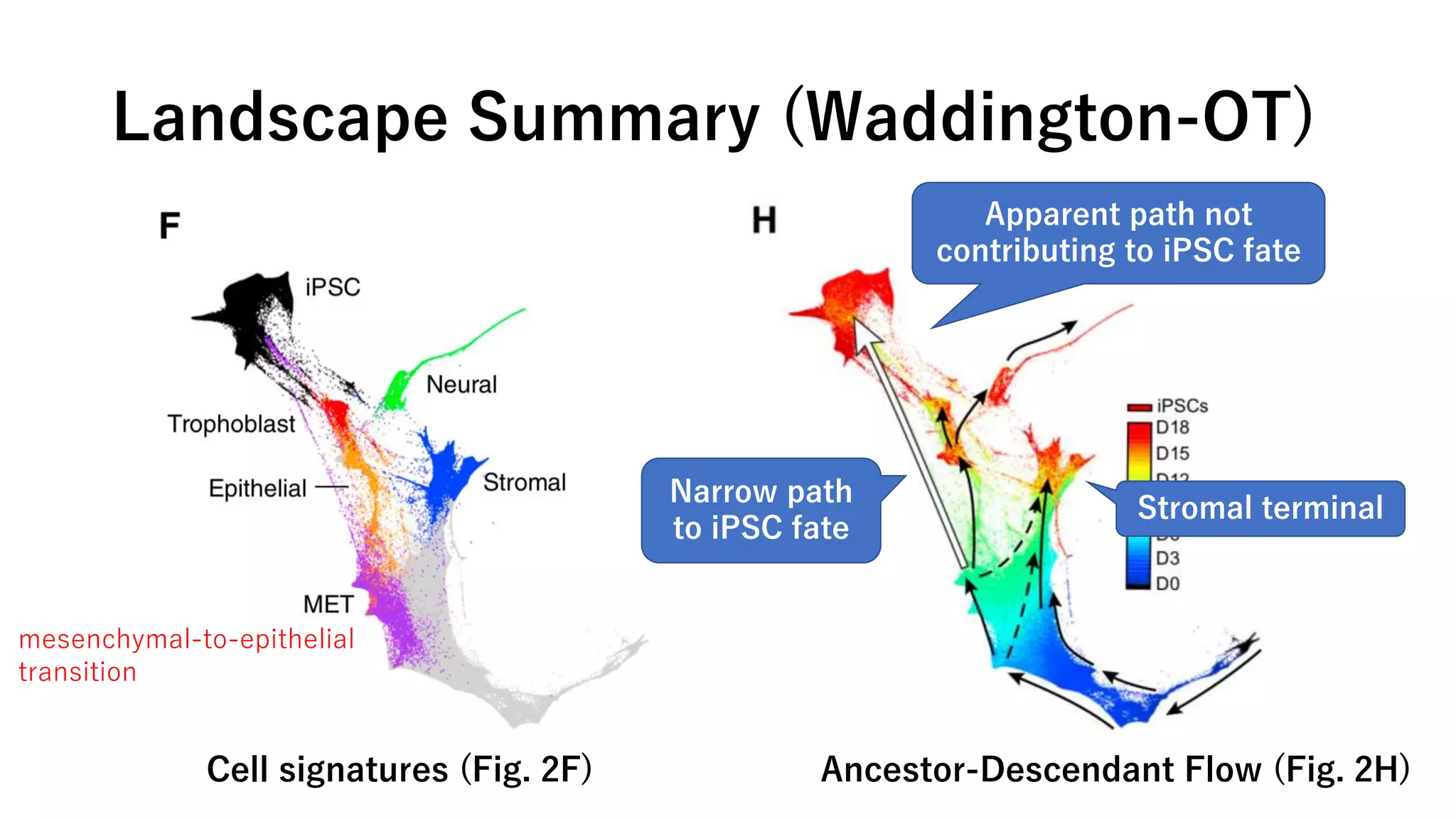
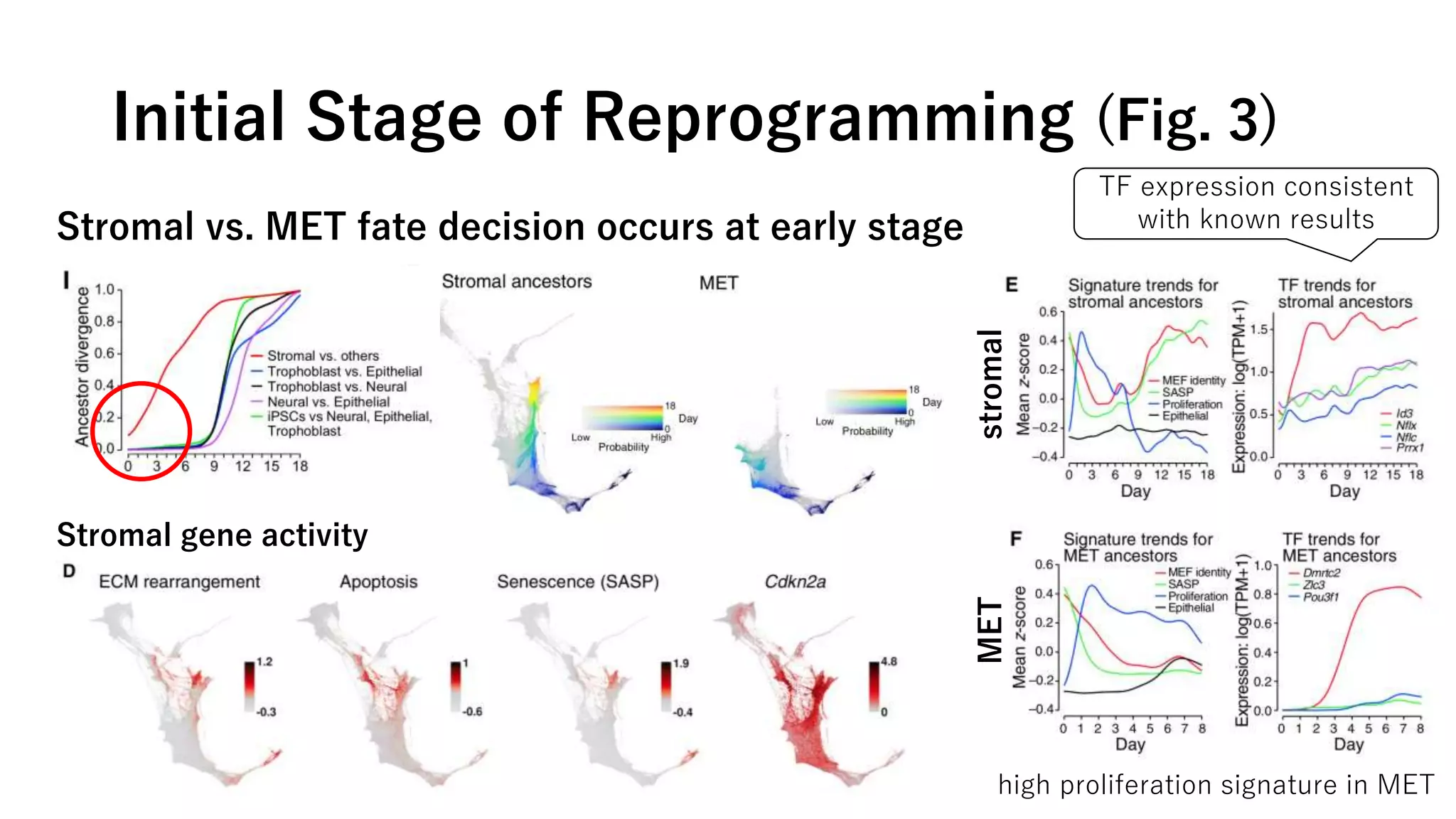
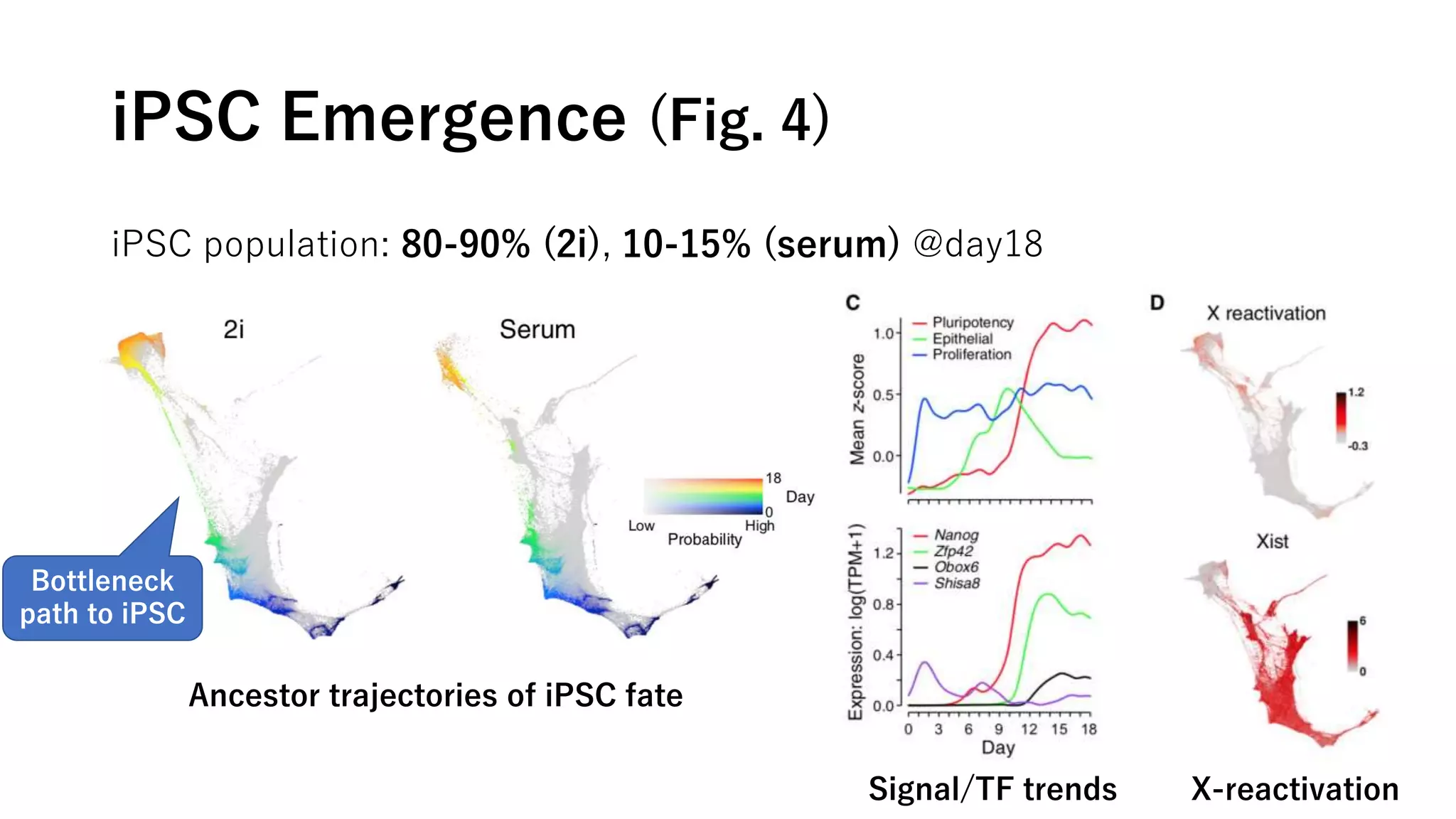
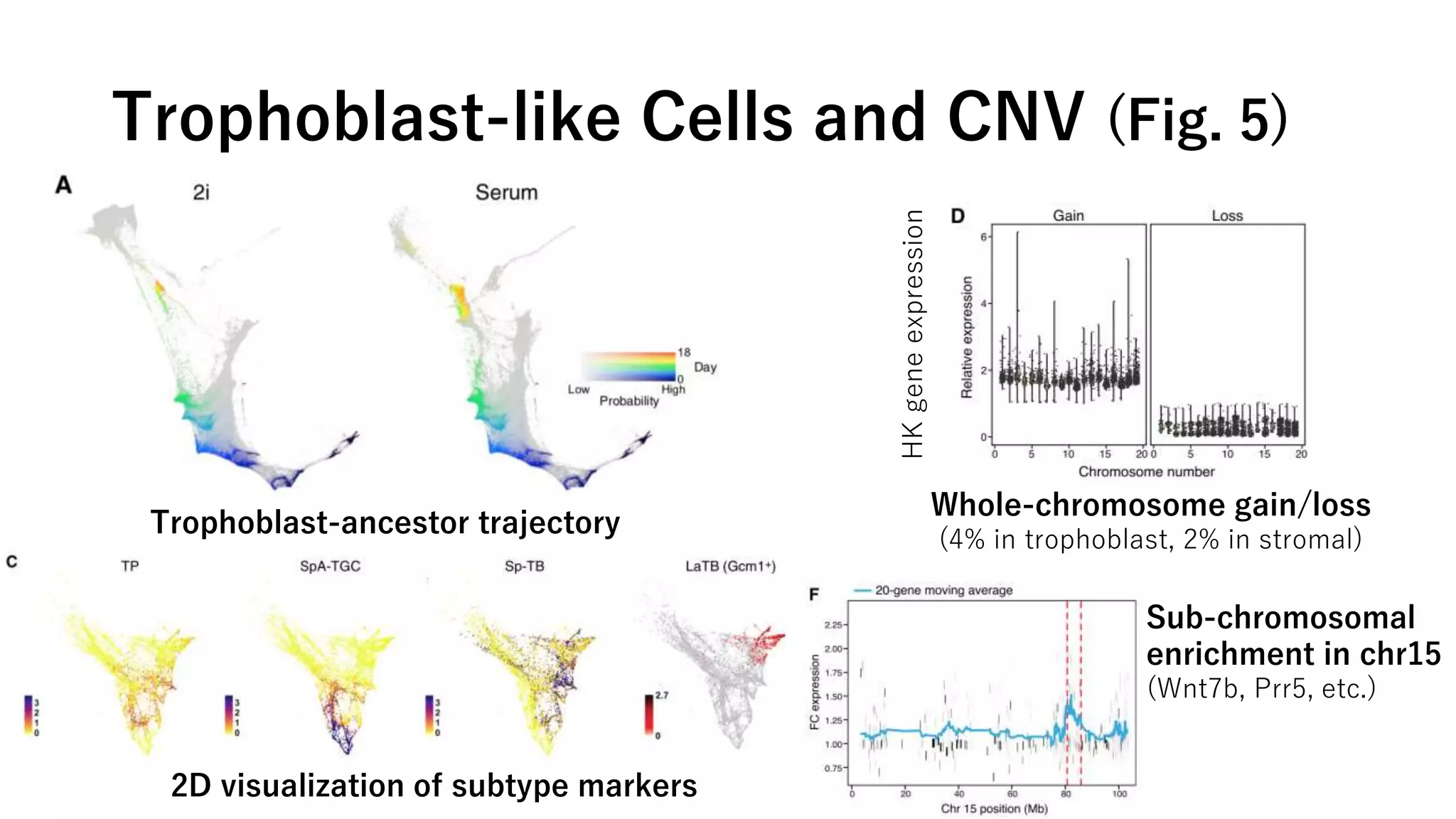
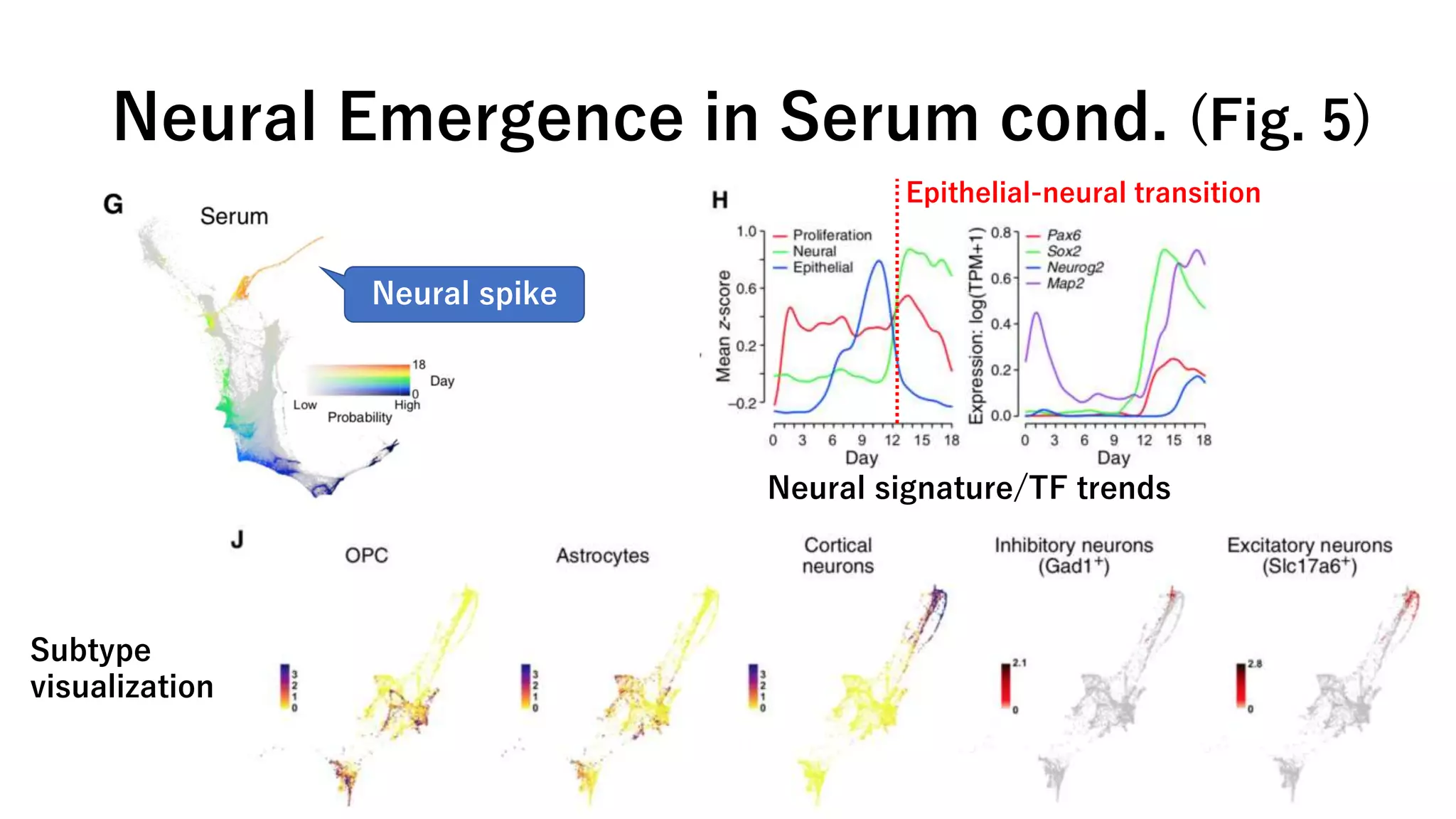
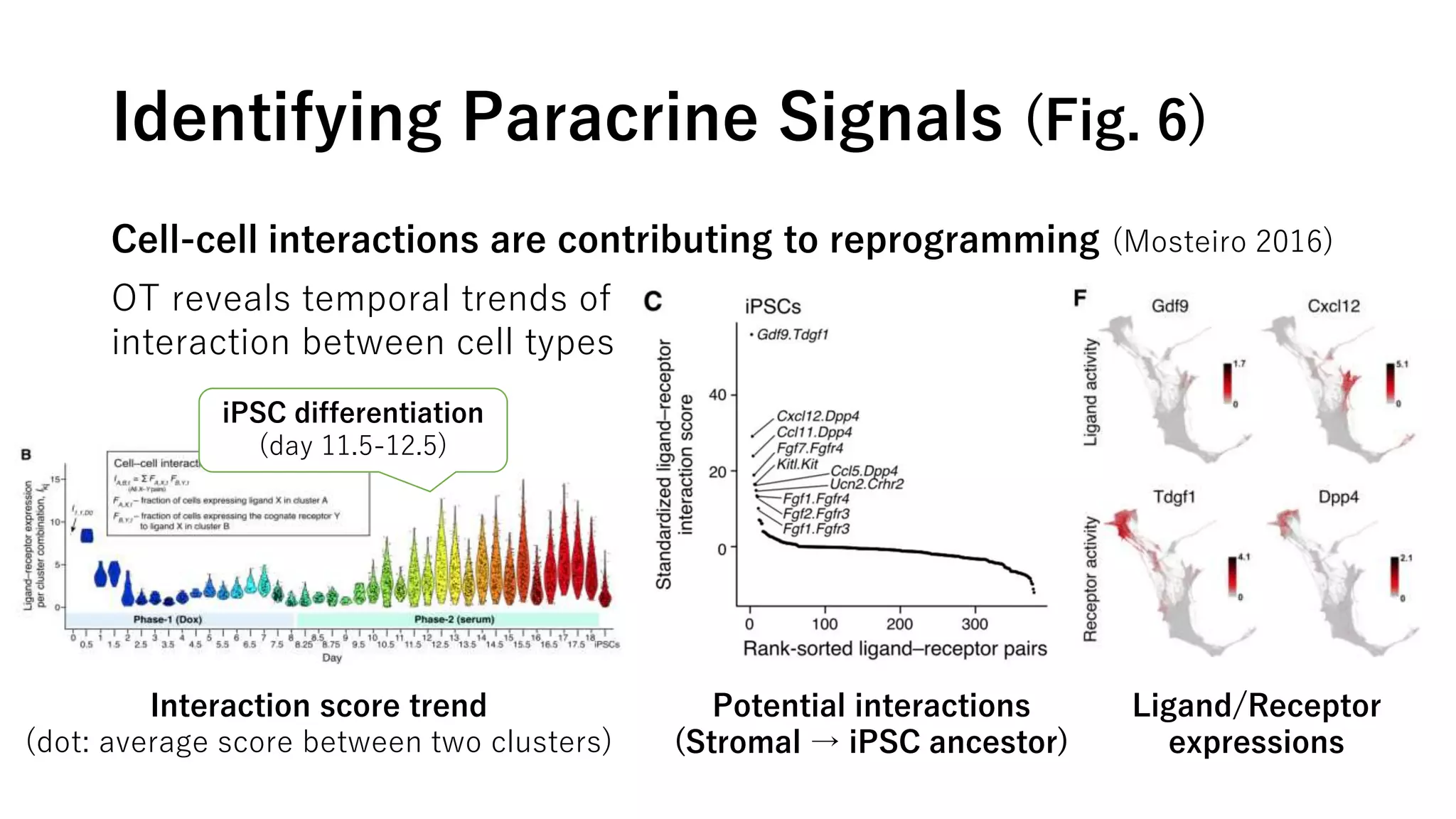
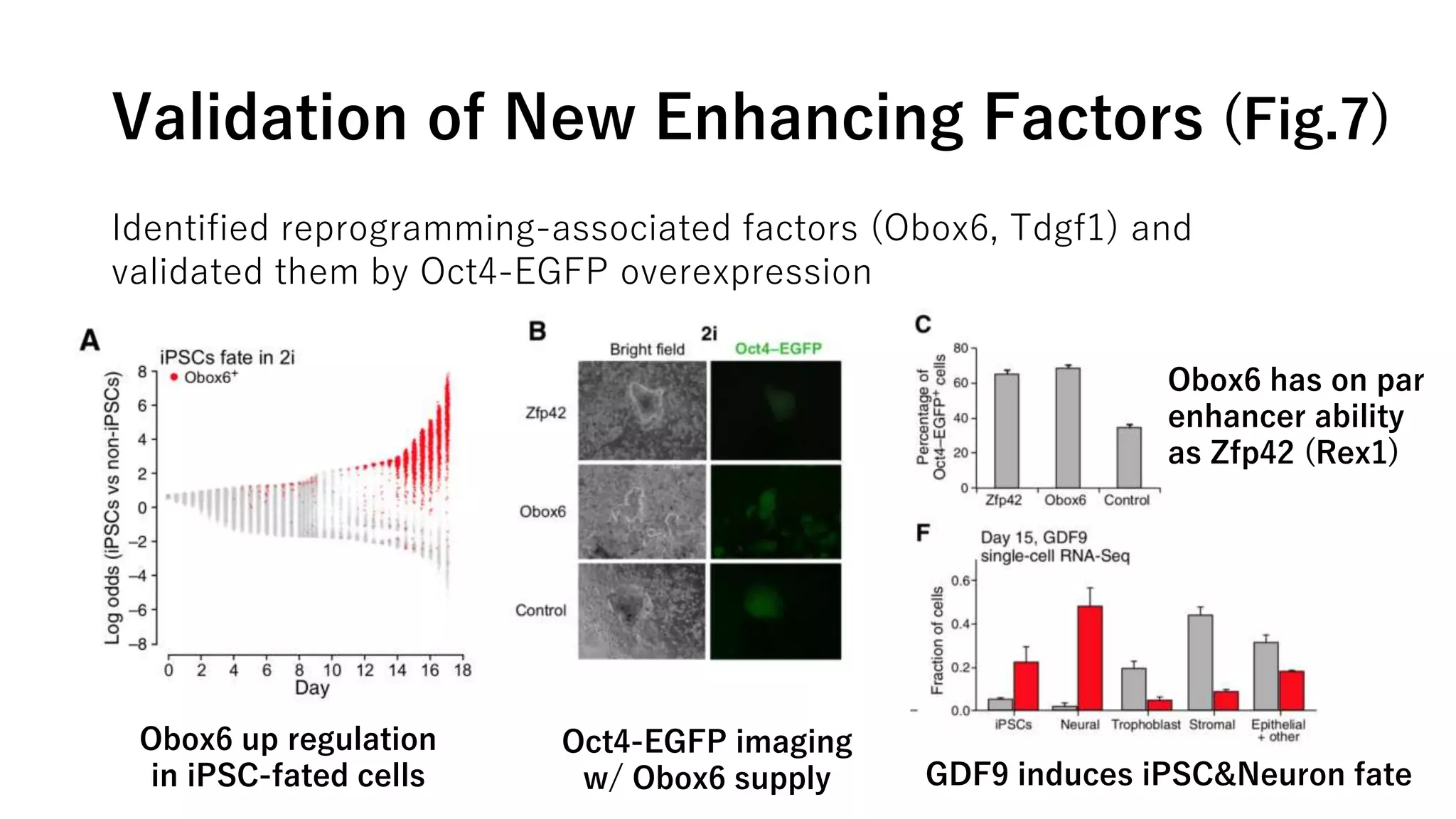
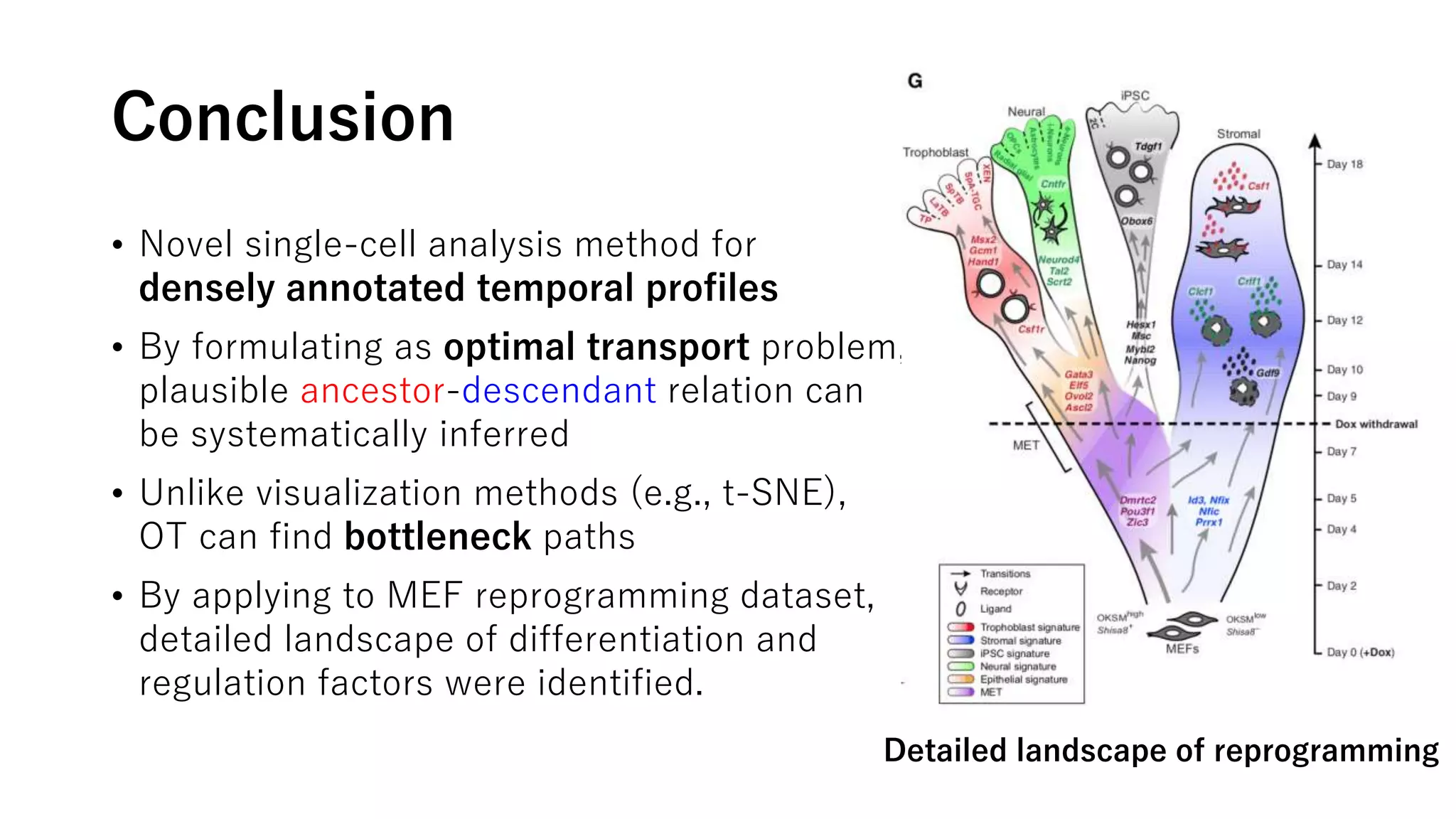
This document summarizes a study that uses single-cell RNA sequencing data from mouse embryonic fibroblast cells undergoing reprogramming to induced pluripotent stem cells over 18 days. The study develops a new computational method called Waddington-optimal transport that models cell differentiation as an optimal transport problem to infer developmental trajectories and ancestor-descendant relationships between cells across time points. Applying this method to the reprogramming data revealed multiple cell fates including induced pluripotent stem cells, stromal cells, and trophoblast-like cells. It also identified transcription factors and signaling pathways important for reprogramming. The method provides a novel way to systematically analyze densely sampled temporal single-cell gene expression data.