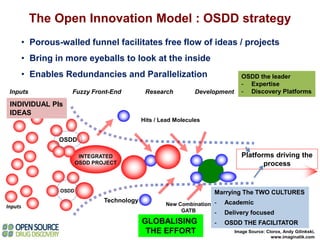
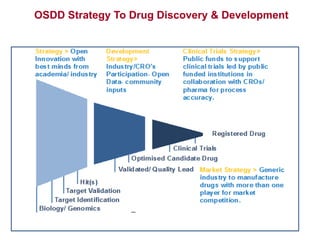
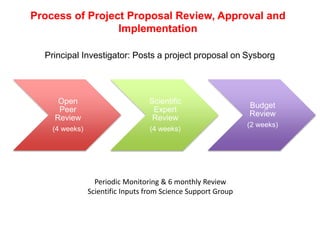
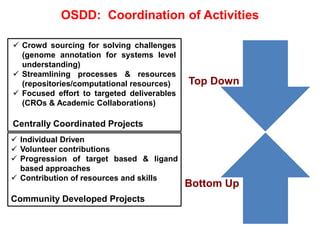
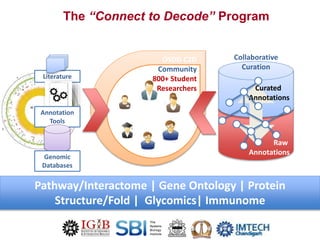
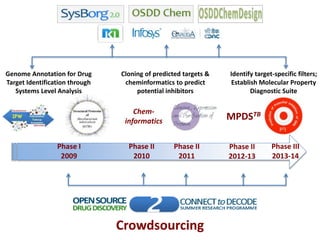
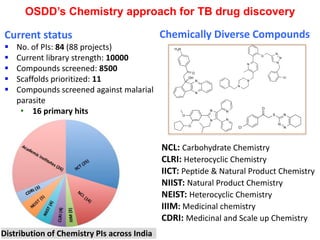
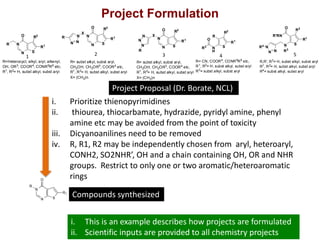
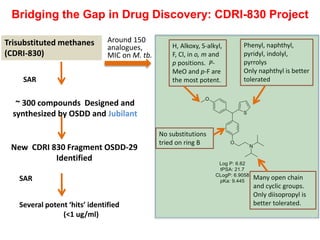
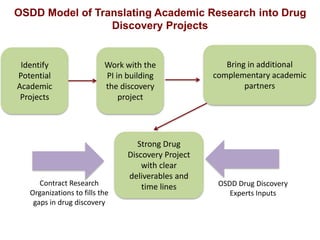
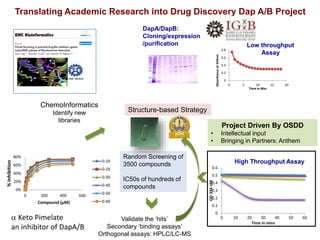
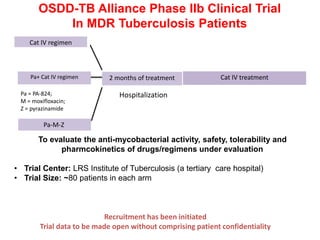
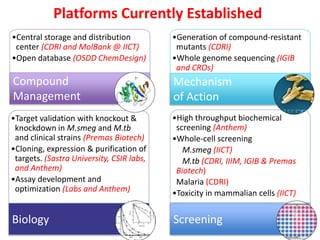
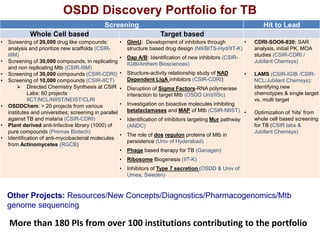
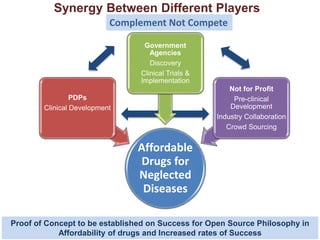
The Open Source Drug Discovery (OSDD) strategy uses an open innovation model with a porous-walled funnel to facilitate the free flow of ideas and projects. It brings in more contributors to look at projects and enables redundancies and parallelization. OSDD acts as a facilitator to marry academic and delivery-focused approaches and provides expertise, discovery platforms, and coordination of activities from both individual and centrally coordinated projects. OSDD has established multiple platforms for drug discovery including compound management, screening, target validation, and mechanistic studies. It has an extensive portfolio involving over 180 principal investigators from over 100 institutions working on projects ranging from whole cell screening to structure-based drug design.