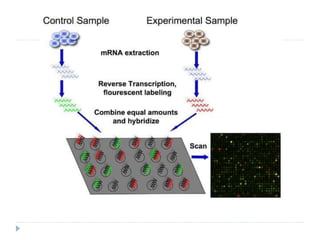
The document discusses nucleic acid microarrays, which are laboratory tools used to analyze gene expression across thousands of genes simultaneously. It covers the history, principles, types of DNA microarrays, and their applications in gene expression analysis, disease diagnosis, drug discovery, and toxicological research. The core techniques involve hybridization, fluorescence detection, and specific microarray types like cDNA and SNP microarrays.