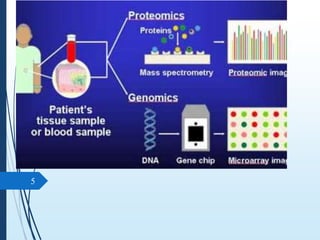
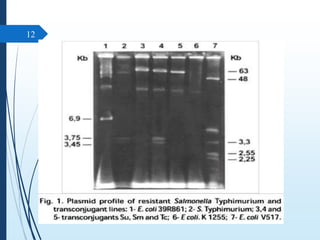
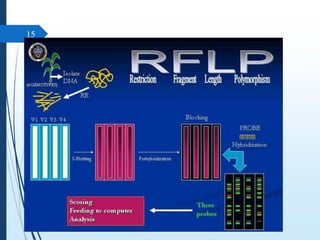
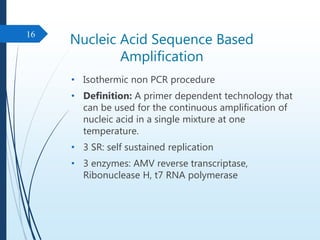
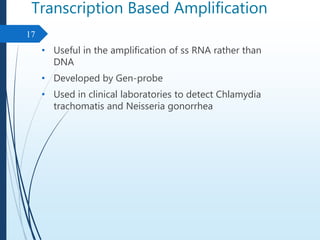
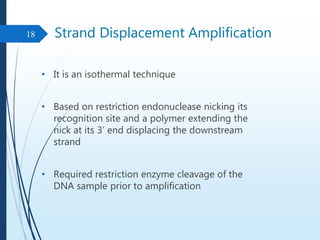
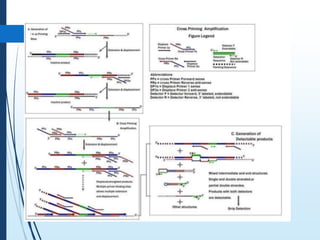
Molecular diagnostics techniques analyze biological markers in the genome and proteome for disease diagnosis and personalized medicine. The history of these techniques includes pivotal developments from early genetic tests for thalassemia to the establishment of professional organizations. Key applications include pathogen detection, antibiotic resistance identification, and forensic uses, with techniques like plasmid profiling, nucleotide sequencing, and amplification methods being critical.