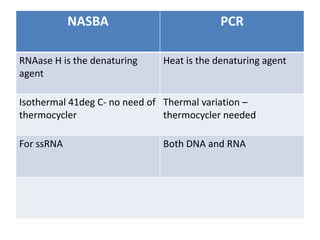
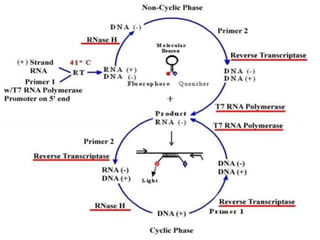
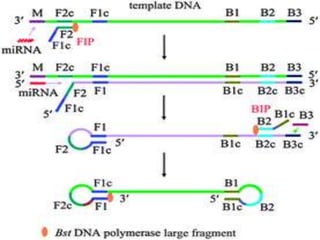
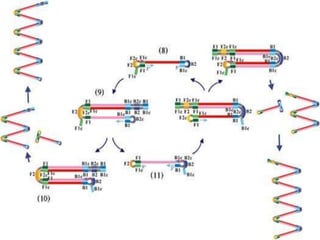
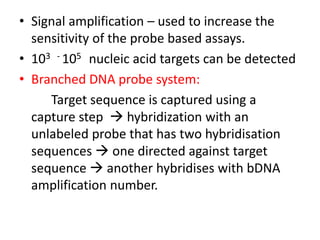
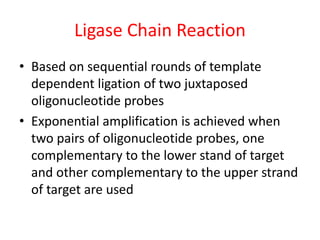
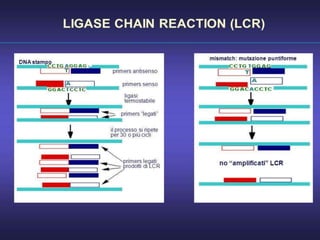
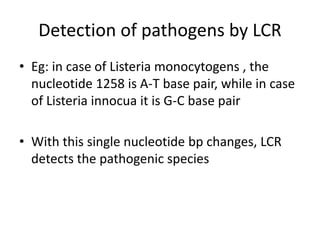
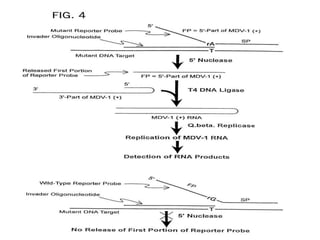
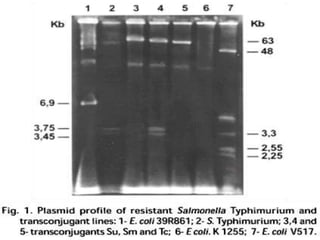
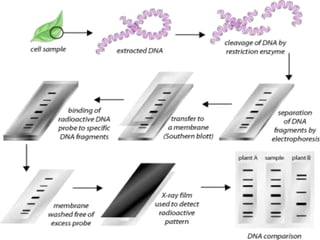
This document discusses various molecular techniques used for diagnosis of infectious diseases. It notes that molecular methods are most useful for pathogens that are difficult to detect by conventional methods, like Mycobacterium tuberculosis and Chlamydia trachomatis. It describes techniques like PCR, NASBA, TBA, SDA, LAMP that amplify nucleic acids from pathogens. Other methods discussed include plasmid profiling, nucleotide sequencing, restriction fragment length polymorphism (RFLP), and nucleic acid hybridization. The document provides details on how several of these techniques work and their applications in microbial identification, detection of antibiotic resistance, and epidemiological studies.