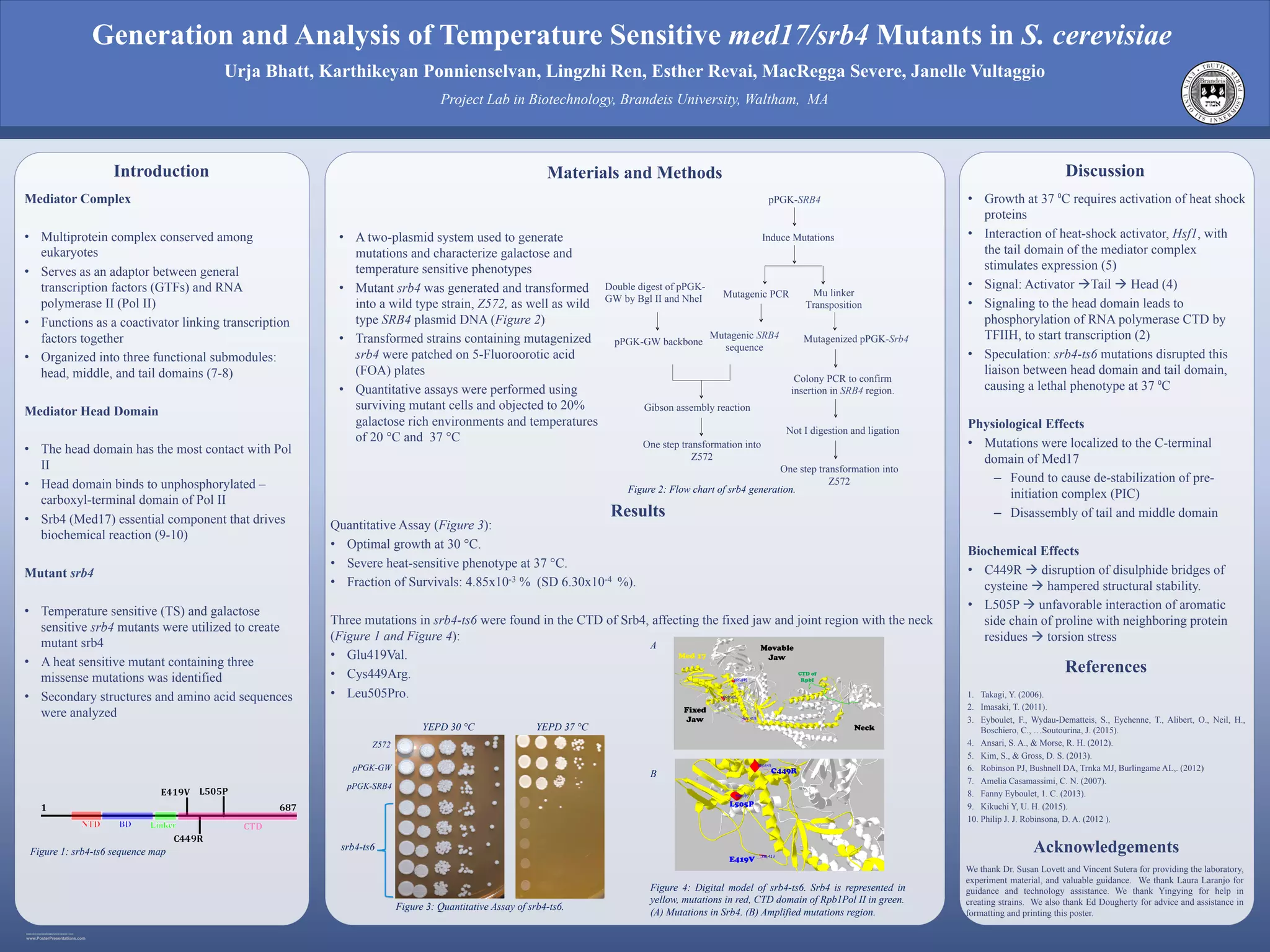
1) Researchers generated temperature sensitive mutations in the MED17/SRB4 gene in Saccharomyces cerevisiae and identified one mutant, srb4-ts6, that was heat sensitive.
2) Quantitative assays found srb4-ts6 grew optimally at 30°C but had severe growth defects at 37°C.
3) DNA sequencing revealed srb4-ts6 contained three missense mutations in the C-terminal domain of Srb4 that are believed to disrupt interactions between the mediator complex's head and tail domains, preventing heat shock gene transcription and causing heat sensitivity.