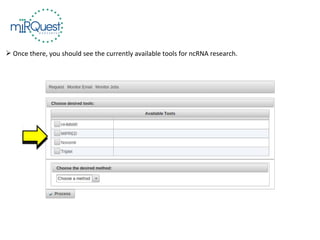
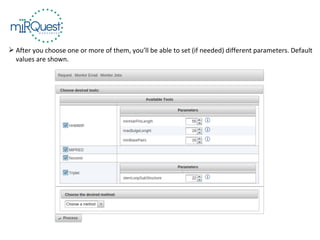
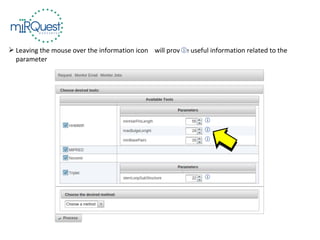
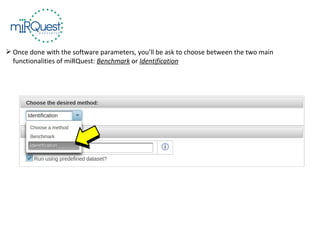
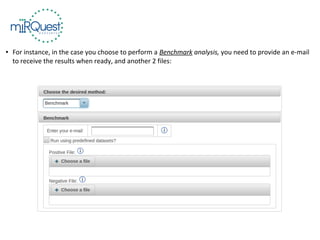
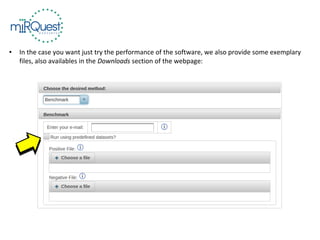
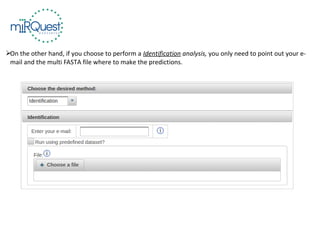
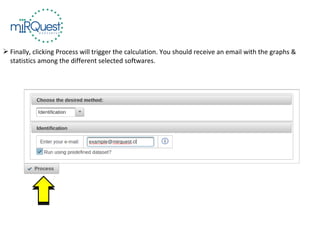
This tutorial explains how to use the miRQuest tool to perform benchmarking and identification of microRNAs (miRNAs). It describes accessing the tool interface and selecting prediction tools, parameters, and file inputs for either benchmarking known miRNA sequences or identifying miRNAs in a user's dataset. Benchmarking requires positive and negative sequence files while identification only needs a multi-FASTA file. Process triggers the analysis and results are emailed.