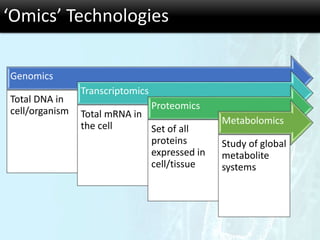
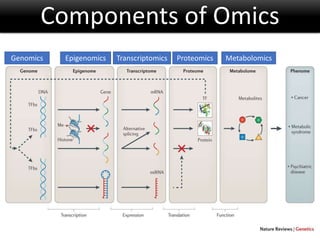
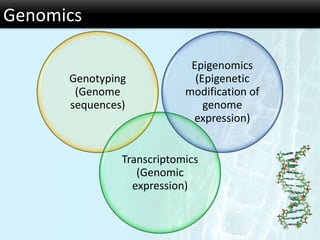
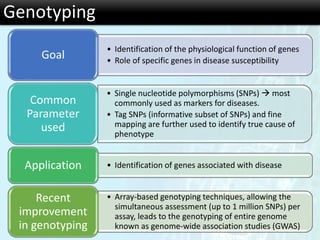
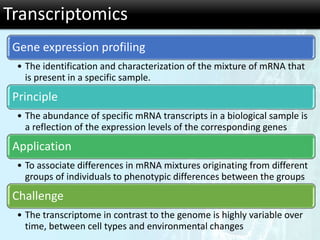
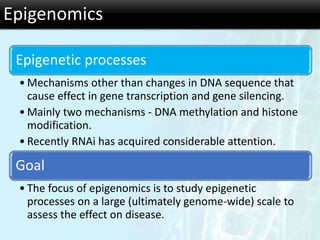
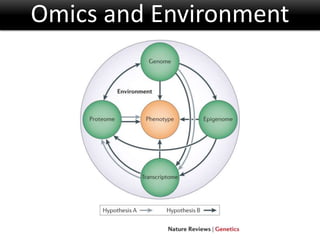
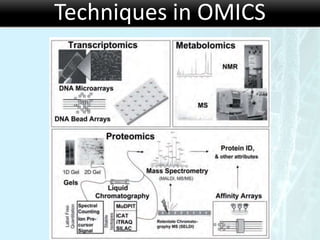
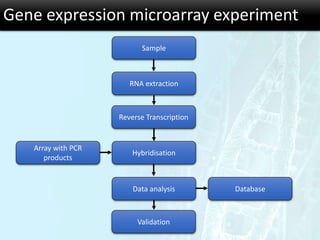
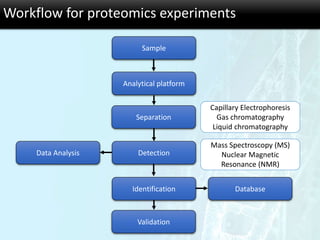
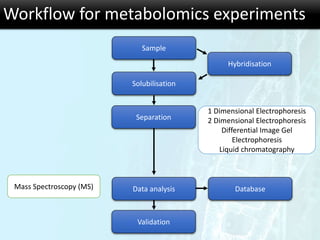
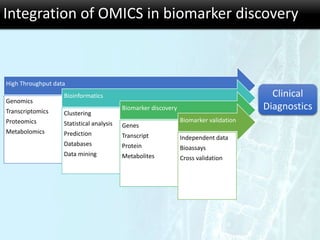
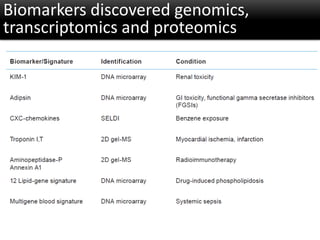
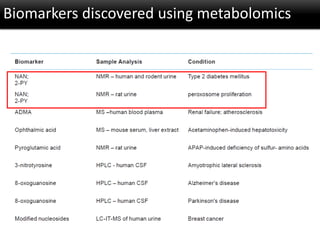
The document discusses the foundational concepts of various 'omics' fields including genomics, transcriptomics, proteomics, and metabolomics, highlighting their historical development and technological advancements. It outlines the methodologies used in each field, their goals, applications, and challenges, as well as the integration of these technologies in biomarker discovery. Overall, 'omic' technologies enable comprehensive analysis of biological samples, with mass spectrometry being a key tool in proteomic and metabolomic research.