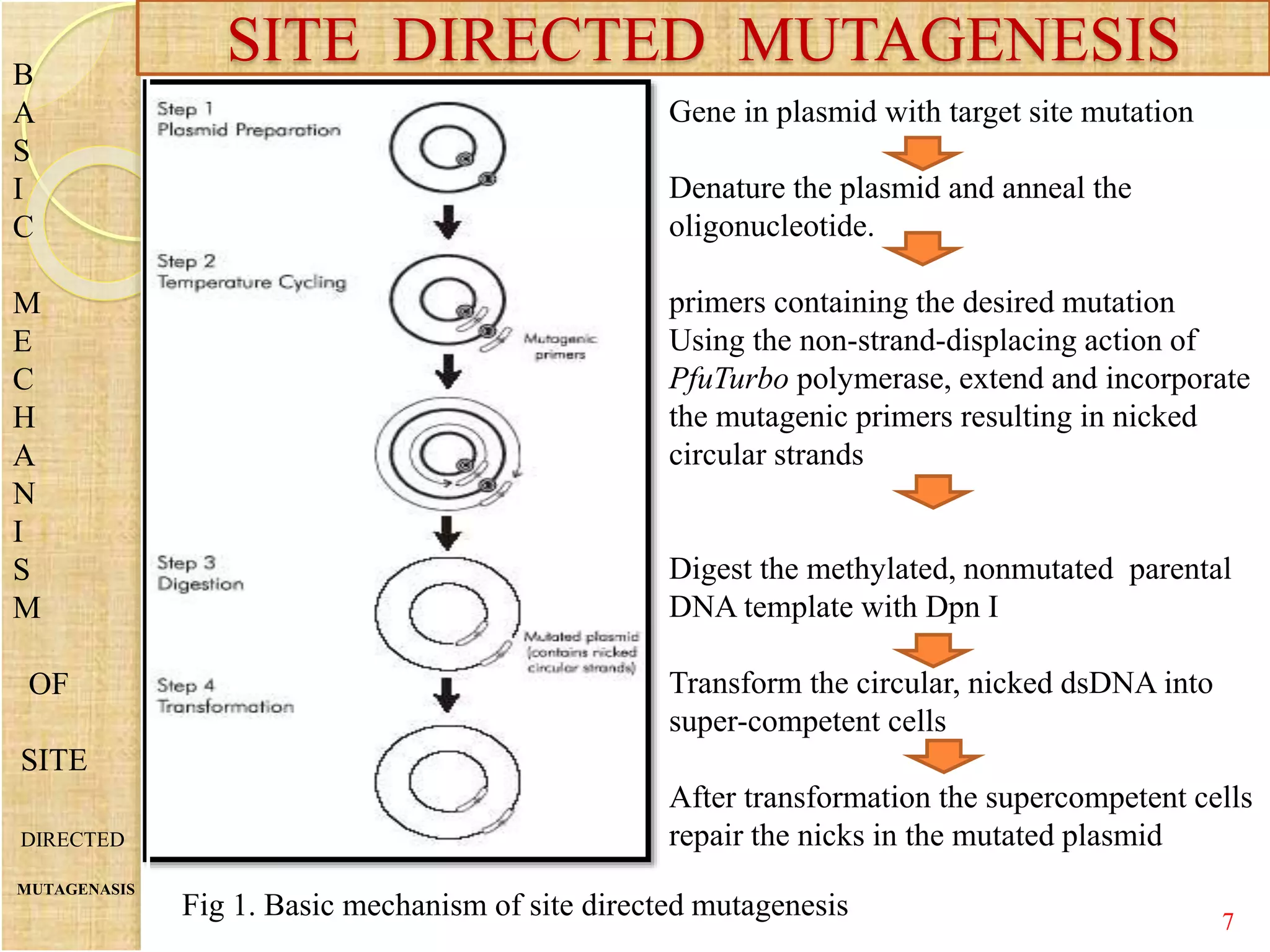
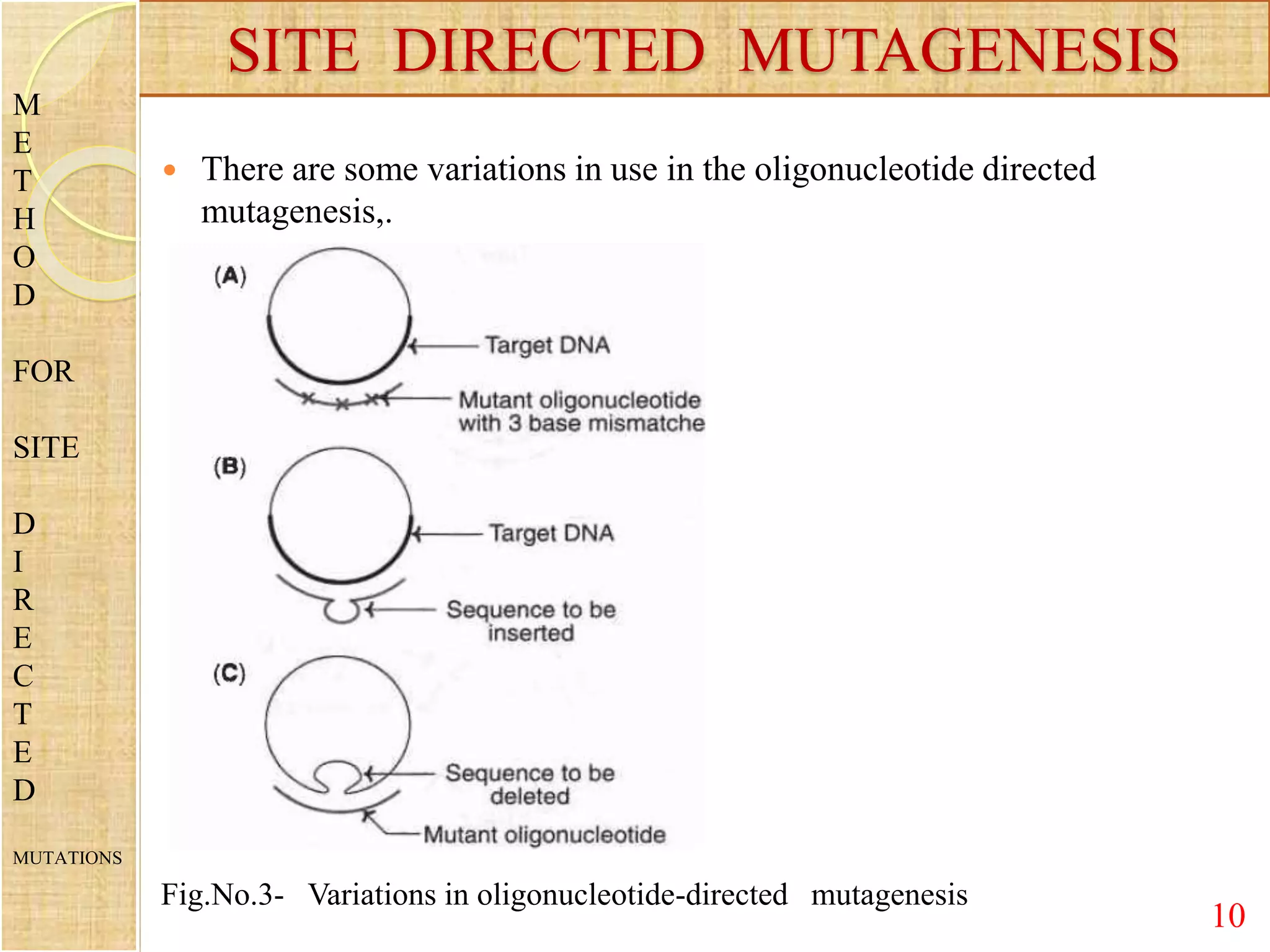
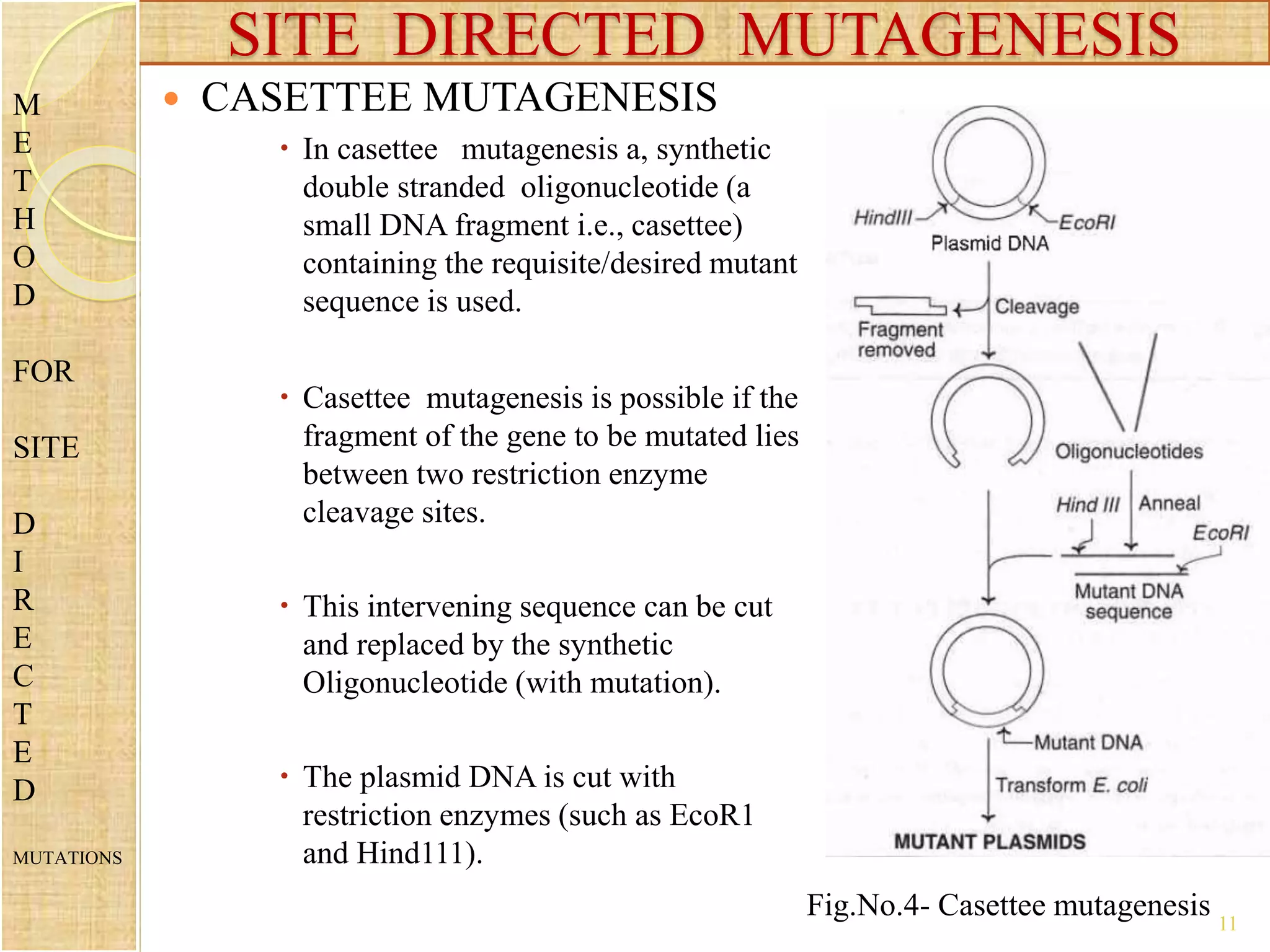
Site-directed mutagenesis is a technique used to generate specific mutations in DNA at predetermined locations. It involves using a synthetic oligonucleotide primer containing the desired mutation to introduce changes into the DNA sequence during in vitro DNA replication or PCR. This allows researchers to study the effects of mutations and engineer proteins with improved or customized properties. Common methods for site-directed mutagenesis include using single or double primers, cassette mutagenesis by replacing DNA fragments, and PCR-based mutagenesis. The technique has various applications in investigating protein function and developing proteins for commercial uses.