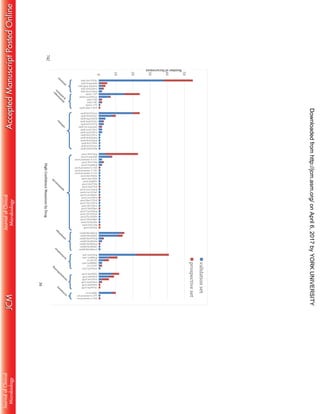
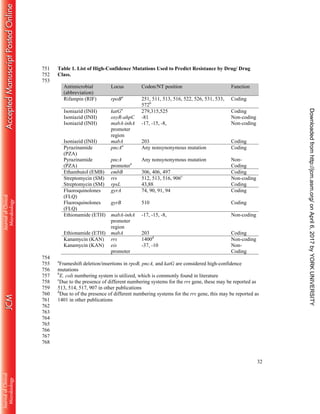
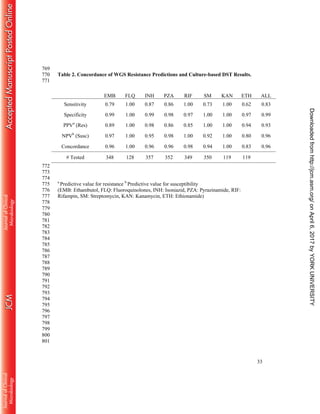
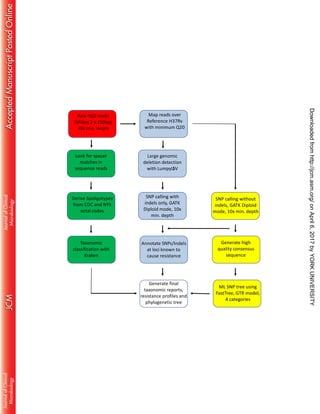
This document describes the validation of a whole genome sequencing (WGS) assay implemented in a public health laboratory to characterize Mycobacterium tuberculosis complex strains. The assay provides rapid identification and comprehensive drug resistance profiles for eight drugs. Validation using 608 clinical isolates found 99% accurate identification and 96% concordance between WGS and culture-based drug susceptibility testing. WGS resistance profiles are reported an average of 9 days sooner for first-line drugs and 32 days sooner for second-line drugs compared to culture-based methods.
























![25
Mycobacterium tuberculosis Isolates with Specific rpoB Mutations. J Clin Microbiol603
51(8):2641-2645. https://doi.org/10.1128/JCM.02741-12.604
30.Eilertson B, Maruri F, Blackman A, Herrera M, Samuels DC, Sterling TR. 2014. High605
Proportion of Heteroresistance in gyrA and gyrB in Fluoroquinolone-Resistant606
Mycobacterium tuberculosis Clinical Isolates. Antimicrob Agents Chemother607
58(6):3270-3275. https://doi.org/10.1128/AAC.02066-13.608
31.Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, Balloux F. 2015.609
Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis610
outbreak strain. Nat Commun 6:7119. https://doi.org/10.1038/ncomms8119.611
32.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser JM. 1997. Analysis of the oxyR-ahpC612
region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex613
organisms recovered from diseased humans and animals in diverse localities. Antimicrob614
Agents Chemother 41(3):600-606.615
33.Feuerriegel S, Oberhauser B, George AG, Dafae F, Richter E, Rüsch-Gerdes S, Niemann616
S. 2012. Sequence analysis for detection of first-line drug resistance in Mycobacterium617
tuberculosis strains from a high-incidence setting. BMC Microbiol 12:90.618
https://doi.org/10.1186/1471-2180-12-90.619
34.Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN,620
Chatterjee D, Fleischmann R, Alland D. 2013. Evolution of high-level ethambutol-621
resistant tuberculosis through interacting mutations in decaprenylphosphoryl-[beta]-D-622
arabinose biosynthetic and utilization pathway genes. Nat Genet 45:1190–1197. https://623
doi.org:10.1038/ng.2743.624
35.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification625
using exact alignments. Genome Biol 15(3):R46. https://doi.org/10.1186/gb-2014-15-3-626
r46.627
36.Thapa J, Nakajima C, Maharjan B, Poudel A, Suzuki Y. 2015. Molecular characterization628
of Mycobacterium orygis isolates from wild animals of Nepal. Jpn J Vet Res 63:151–8.629
https://doi.org/10.14943/jjvr.63.3.151630
37.Rodriguez-Campos S, Smith NH, Boniotti MB, Aranaz A. 2014. Overview and631
phylogeny of Mycobacterium tuberculosis complex organisms: Implications for632
diagnostics and legislation of bovine tuberculosis. Res Vet Sci 97(Suppl):S5–S19.633
38.Feuerriegel S, Köser CU, Niemann S. 2014. Phylogenetic polymorphisms in antibiotic634
resistance genes of the Mycobacterium tuberculosis complex. J Antimicrob Chemother635
69(5):1205–10. https://doi.org/10.1093/jac/dkt535.636
39.García-Sierra N, Lacoma A, Prat C, Haba L, Maldonado J, Ruiz-Manzano J, Gavin P,637
Samper S, Ausina V, Domínguez J. 2011. Pyrosequencing for Rapid Molecular Detection638
of Rifampin and Isoniazid Resistance in Mycobacterium tuberculosis Strains and Clinical639
Specimens. J Clin Microbiol 49(10):3683-3686. https://doi.org/10.1128/JCM.01239-11.640
40.Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. 2003. Mutations in the rpoB Gene of641
Multidrug-Resistant Mycobacterium tuberculosis Isolates from China. J Clin Microbiol642
41(5):2209-2212. https://doi.org/10.1128/JCM.41.5.2209-2212.2003.643
41.Mani C, Selvakumar N, Narayanan S, Narayanan PR. 2001. Mutations in the rpoB Gene644
of Multidrug-Resistant Mycobacterium tuberculosis Clinical Isolates from India. J Clin645
Microbiol 39(8):2987-2990. https://doi.org/10.1128/JCM.39.8.2987-2990.2001.646
42.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT,647
Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L,648
onApril6,2017byYORKUNIVERSITYhttp://jcm.asm.org/Downloadedfrom](https://image.slidesharecdn.com/10-170831202423/85/10-1128-jcm-00298-17-25-320.jpg)