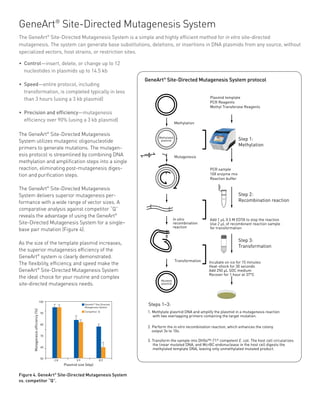
The GeneArt® Seamless Cloning and Assembly Kit enables the simultaneous and directional cloning of 1 to 4 PCR fragments, consisting of any sequence, into any linearized vector, in a single 30-minute room temperature reaction. The kit contains everything required for the assembly of DNA fragments, and their transformation into E. coli for selection and growth of recombinant vectors.
• Speed and Ease — Clone up to 4 DNA fragments, with sequence of your choice, simultaneously in a single vector (up to 13 Kb); no restriction digestion, ligation or recombination sites required
• Precision and Efficiency —Designed to let you clone what you want, where you want, in the orientation you want, and achieve up to 90% correct clones with no extra sequences left behind
• Vector Flexibility — Use our linear vector or a vector of your choice
• Free Tools — Design DNA oligos and more with our free web-based interface that walks you step-by-step through your project
• Diverse Applications — Streamline many synthetic biology and molecular biology techniques through the rapid combination, addition, deletion, or exchange of DNA segments
For cloning more than 4 DNA fragments, final molecules larger than 110 Kb, or for using pre-existing DNA elements too long to be amplified by PCR; please consider the GeneArt® High-Order Genetic Assembly System (cat# A13285).
Simple and Fast Clone Creation
GeneArt® Seamless Cloning is a simple, two step process, consisting of a tube based assembly reaction followed by transformation into One Shot® Chemically Competent TOP10 E. coli. The kit uses the properties of a proprietary enzymatic mix to assemble DNA fragments with shared terminal end homology without leaving any extra sequences or scars behind (seamless). A portion of the assembly reaction is then transformed into the provided competent TOP10 cells, yielding clones ready for analysis the next day. The required 15-base pair end-homology can be easily engineered by PCR-amplification with custom DNA oligos.
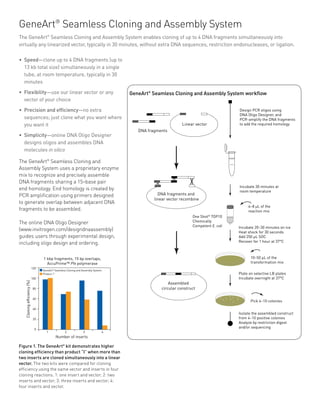
Cloning Efficiency, Flexibility, and Precision
With the GeneArt® Seamless Cloning and Assembly Kit the main factors effecting cloning efficiency are the size of the DNA elements (100 bp to 5 Kb), the total size of the final molecule (≤ 13 Kb), and the quality and specificity of the fragment. The terminal end of the PCR fragments (A-overhangs or blunt), does not affect the cloning efficiency.
Typical cloning efficiencies for different numbers of fragments cloned into pUC19L are the following:
• 90% for a single 5 Kb DNA element
• 70% for 4 fragments of 1 Kb each
• 40% for 4 DNA fragments of 2 Kb each
For more information visit:
https://products.invitrogen.com/ivgn/product/A13288?CID=cloningkit-SS-12312

![GeneArt® High-Order Genetic Assembly System
The geneArt® High-order genetic Assembly system is a highly efficient, vector-independent system for the simultaneous
and seamless assembly of up to 10 DNA fragments (preexisting or synthetic) in vivo, totaling up to 110 kb in construct size.
• Speed—clone 10 or more DNA fragments 100
simultaneously into a single vector (up to 110 kb
80
total); assemble existing DNA fragments without
Cloning efficiency (%)
restriction digestion or pcR amplification 60
• Flexibility—use our linear vector or a vector of your 40
choice; oligonucleotide stitching feature allows
end-editing and reuse of preexisting DNA fragments 20
without the need for reamplification 0
1 3 10
• Precision and efficiency—no extra sequences; just Number of 10 kb DNA fragments
clone what you want where you want it Figure 3. Cloning efficiency of the GeneArt® High-Order
Genetic Assembly System with increasing numbers of 10
• Simplicity—online DNA oligo Designer walks you kb PCR fragments. Even as 100 kb of PCR fragments are
through your project step by step; design your DNA assembled, the cloning efficiency remains over 65%, proving
the system is a reliable solution for assembly of complex DNA
oligos and assemble your DNA molecule in silico
molecules.
(www.invitrogen.com/designdnaassembly)
The GeneArt® High-Order Genetic
Assembly System relies on yeast’s
ability to take up and recombine A Fragment Oligo Cloning
DNA fragments with high efficiency size size efficiency
via transformation-associated
recombination, greatly reducing in x x x x
1 x 1 kb 60-mer 94%
x x x x
vitro handling of DNA and eliminating
the need for restriction digestion and x x x x x x
2 x 5 kb 80-mer 75%
ligation. x x x x x x
In cases where DNA fragments do x x x x x x x x [3 x 5 kb] + [2 x 0.5 kb] 60-mer 37%
not share end homology (i.e., existing x x x x
[3 x 5 kb] + [2 x 0.5 kb] 80-mer 75%
DNA fragments not created by PCR),
they can be “stitched” together using
B
recombinant linkers that provide 80-mer
2 x 5 kb 63%
sequence overlaps to promote
v
x x x x x x 10 bp insertion
recombinatorial joining of unrelated x x x
v
x x x
2 x 5 kb 80-mer 50%
DNA fragments (Figure 2A). 20 bp insertion
x x x x x x 80-mer
In addition, oligonucleotide stitch- x x x x x x
2 x 5 kb
12 bp insertion
87%
ing allows editing of the fragment
junctions to generate deletions and Figure 2. Oligonucleotide stitching. (A) Oligonucleotide stitching enables joining
insertions (Figure 2B). This feature of unrelated DNA fragments. (B) Oligonucleotide stitching can be used for junction
also allows the reuse of existing DNA editing.
fragments or elements obtained by
restriction enzyme digestion without
the need to reamplify them by PCR.](https://image.slidesharecdn.com/geneartkitbrochure-120123123906-phpapp01/85/GeneArt-Seamless-Cloning-and-Assembly-Kit-5-320.jpg)