table6vert
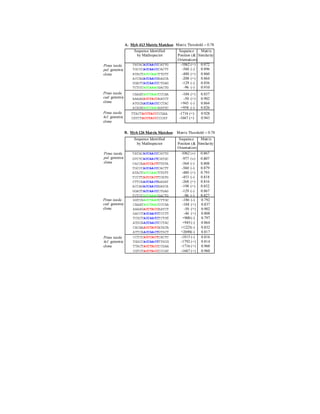
- 1. Pinus taeda pal: genomic clone Pinus taeda 4cl: genomic clone Pinus taeda cad: genomic clone TTACTACCTACCCCGAA -1716 (+) 0.928 CGTCTACCTACCCCCAT -1667 (+) 0.943 Sequence Identified Sequence Matrix by MatInspector Position (& Similarity Orientation) TATACACCAACCCATTG -1062 (+) 0.872 TGCCCACCAACCCACTT -560 (-) 0.896 ATACTACCCAACTTGTT -480 (+) 0.860 ACCAGACCAACCAAGCA -208 (+) 0.864 GGATTACCAACCCTGAG -129 (-) 0.856 TCTCCACCAAACGACTG -96 (-) 0.910 CAAATACCTAACCCCAA -104 (+) 0.837 AAAAGACCTACCAATCT -50 (+) 0.902 ATGCAACCAACCCCTAC +945 (-) 0.864 ACAGGACCCAACAATGC +958 (-) 0.826 A. Myb 413 Matrix Matches: Matrix Threshold = 0.78 CCTCCACCCACTCACTC -1815 (-) 0.816 TGGCCACCAACTTTGCG -1792 (+) 0.814 TTACTACCTACCCCGAA -1716 (+) 0.960 CGTCTACCTACCCCCAT -1667 (+) 0.960 B. Myb 126 Matrix Matches: Matrix Threshold = 0.78 Sequence Identified Sequence Matrix by MatInspector Position (& Similarity Orientation) TATACACCAACCCATTG -1062 (+) 0.867 GTCTCACCAACTCATGC -977 (+) 0.807 CACCAACCCACTTTGTA -564 (-) 0.808 TGCCCACCAACCCACTT -560 (-) 0.879 ATACTACCCAACTTGTT -480 (+) 0.793 TCCTTACCCACTTCATG -453 (-) 0.818 CTTCAACCAACTAAGAT -268 (+) 0.816 ACCAGACCAACCAAGCA -198 (+) 0.852 GGATTACCAACCCTGAG -129 (-) 0.867 TCTCCACCAAACGACTG -96 (-) 0.827 GGTCGACCTAATCTTGC -186 (-) 0.792 CAAATACCTAACCCCAA -104 (+) 0.837 AAAAGACCTACCAATCT -50 (+) 0.902 GACCTACCAATCTCCTT -46 (+) 0.808 TCGCTACCAATCTCTGT +900 (-) 0.797 ATGCAACCAACCCCTAC +945 (-) 0.864 CACAAACCTATCATATA +1225(-) 0.832 ATTCAACCAACTGTGCT +2690(-) 0.817 Pinus taeda pal: genomic clone Pinus taeda cad: genomic clone Pinus taeda 4cl: genomic clone