seq alignment.ppt
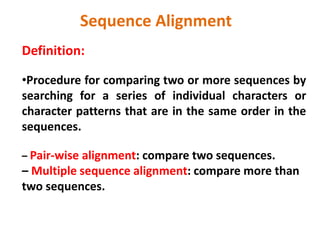
- 1. Definition: •Procedure for comparing two or more sequences by searching for a series of individual characters or character patterns that are in the same order in the sequences. – Pair-wise alignment: compare two sequences. – Multiple sequence alignment: compare more than two sequences. Sequence Alignment
- 2. Need • To find whether two (or more) genes or proteins are evolutionarily related to each Other. • To find structurally or functionally similar regions within proteins.
- 3. • Align abcdef with abdgf • Write second sequence below the first abcdef abdgf • Move sequences to give maximum match between them. • Show characters that match using vertical bar. Example
- 4. Example sequence alignment abcdef || abdgf abcdef || || | ab-dgf
- 5. Matching Similarity vs. Identity • Alignments can be based on finding only identical characters, or (more commonly) can be based on finding similar characters. • Conserved substitutions, semi-conserved substitution, and non conserved substitution are the terms used to define similarity.
- 6. Matching Similarity vs. Identity Different colours denote different chemical groups of amino acids, i.e. hydrophobic, acidic, etc.
- 7. Inferences • In sequence alignment, the degree of similarity between particular regions in the sequences can be interpreted as a rough measure of how conserved a particular region or sequence motif is among lineages • In conserved regions the order of letters does not change during evolution or changes only slightly, i.e. has mostly conservative substitutions. • Sequence motif = sequence pattern that is widespread and has a biological significance resulting in its conservation through evolution.
- 8. Global vs. Local Alignment – Global alignment algorithms which optimize overall alignment between two sequences. – Local alignment algorithms which seek only relatively conserved pieces of sequence. Alignment stops at the ends of regions of strong similarity. – Favors finding conserved patterns in different pairs of sequences.
- 9. • Global LGPSSKQTGKGS-SRIWDN LN-TKSAGKGAIMRLGDA • Local --------GKG-------- ||| --------GKG--------
- 10. Methods for Pair-wise Alignment • Dot matrix analysis • Dynamic Programming (Needleman- Wunsch, Smith-Waterman algorithms) • Word or k-tuple methods (BLAST and FASTA)
- 11. DOT MATRIX
- 12. Interpretation of Dot Matrices • Regions of similarity appear as diagonal runs of dots. • Interruption in middle of diagonal line indicates insertions or deletions. • Parallel diagonal line within the matrix represents repetitive regions of the sequence.
- 13. Uses • Can use dot matrices to align two proteins or two nucleic acid sequences. • Can use to find amino acid repeats within a protein by comparing a protein sequence to itself. • Used in identifying Nucleic Acids secondary structure detecting self complementarily of the sequence. • Used in comparative genomics by predicting gene order conservation between closely related genomes.
- 14. Limitations • A problem with dot matrices for long sequences is that they can be very noisy due to lots of insignificant matches. • Only a pairwise alignment method not suitable for multiple alignment of sequences. • It lacks statistically rigor in assessing the quality of alignment.
- 15. Solution • By using a window (W)/ tuple. – compare character by character within a window (have to choose window size). – require certain fraction of matches within window in order to display it with a “dot”.
- 17. • Initialisation • Matrix fill (scoring) • Traceback (alignment) Dynamic Programming Approach Steps M= (length of sequence i) N= (Length of sequence ii)
- 19. Scoring • For each position, Mi,j is defined to be the maximum score at position i,j; i.e. Mi,j = MAXIMUM [ Mi-1, j-1 + Si,j (match/mismatch in the diagonal) Mi,j-1 + w (gap in sequence #1), Mi-1,j + w (gap in sequence #2)] • In the following case, Mi-1,j-1 will be red, Mi,j-1 will be green and Mi-1,j will be blue.
- 20. • A simple scoring scheme is assumed where – Si,j = 1 if the residue at position i of sequence #1 is the same as the residue at position j of sequence #2 (match score); otherwise – Si,j = 0 (mismatch score) – w = 0 (gap penalty)
- 21. Matrix Fill Step
- 26. Traceback Step
- 30. Final Alignment G A A T T C A G T T A | | | | | | G G A _ T C _ G _ _ A
- 34. Summary • The NW alignment is over the entire length of two Sequences (the traceback starts from the lower right corner of the traceback matrix, and completes in the upper left cell of this matrix). • The Needleman-Wunsch algorithm works in the same way regardless of the length or complexity of sequences and guarantees to find the best alignment. • The Needleman-Wunsch algorithm is appropriate for finding the best alignment of two sequences which are (i) of the similar length. (ii) similar across their entire lengths.
Editor's Notes
- Conserved amino acid substitution are the replacement of an amino acid residue with another one with similar properties such as aspartate and glutamate. They are both negatively charged. Semi-conserved amino acid substitution replaces one residue with another one that has similar steric conformation but does not share chemical properties like substitution of cysteine for alanine or leucine.