More Related Content
More from GenomeInABottle (20)
Mason u41 grant figures
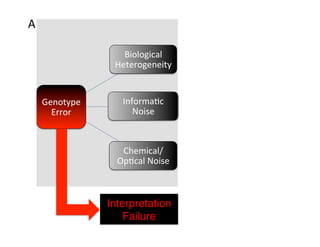
- 3. A
B
5-‐10%
-‐
Non-‐reproducible
Regions
of
the
Genome
~90-‐95%
-‐
Reproducible
Regions
of
the
- 5. Complete !
Genomics!
Overlap! 1000 Genomes!
15,309,617! 13,029,568!12,357,915!
A
B
C
D
E
Region
%(of(
genome
Gold 95.07%
Silver 2.95%
Bronze 0.02%
Lead 1.96%
50x 100x 200x
Gold 100% 100% 100%
Silver 85% 90% 94%
Bronze 54% 68% 82%
Lead 519 6,589 22,164
WGS5Concordance
0%#
1%#
2%#
3%#
Silver# Bronze# Lead#
%"of"Medically"
Relevant"SNVs"
N=32
WGS
of
replicate
DNA
@
30X
coverage
(Rosenfeld,
Mason,
Smith,
2012)
- 6. A)
B)
C)
D)
E)
F)
G)
H)
I)
- 7. RM
Tracking
&
Dissemina5on
• RM
Maintenance
(Salit)
• RM
distribu5on
(Zook)
• RM
Tracking
(Mason)
RM
Tes5ng
and
Sequencing
Sites
• Cornell
(Mason,
Grills)
• MSSM(Schadt,
Sebra)
• ABRF
(Grills,
NARG)
• Vendors
(ILM,
CG,
PAC,
LIF,
BIO)
Informa5cs
and
Genome
Analysis
• GCAT
(Mi_elm
• Arvados
(Zara
• Gmake
(Maso
- 12. 74.74%
0.55%
19.01%
5.60%
0.10%
Popula5on
gene5cs
Protein
structure
Sequence
analysis
Other
Sequence
alignment
0
20
40
60
80
100
120
140
160
2005
2006
2007
2008
2009
2010
2011
2012
Number
of
users
Year
A
B