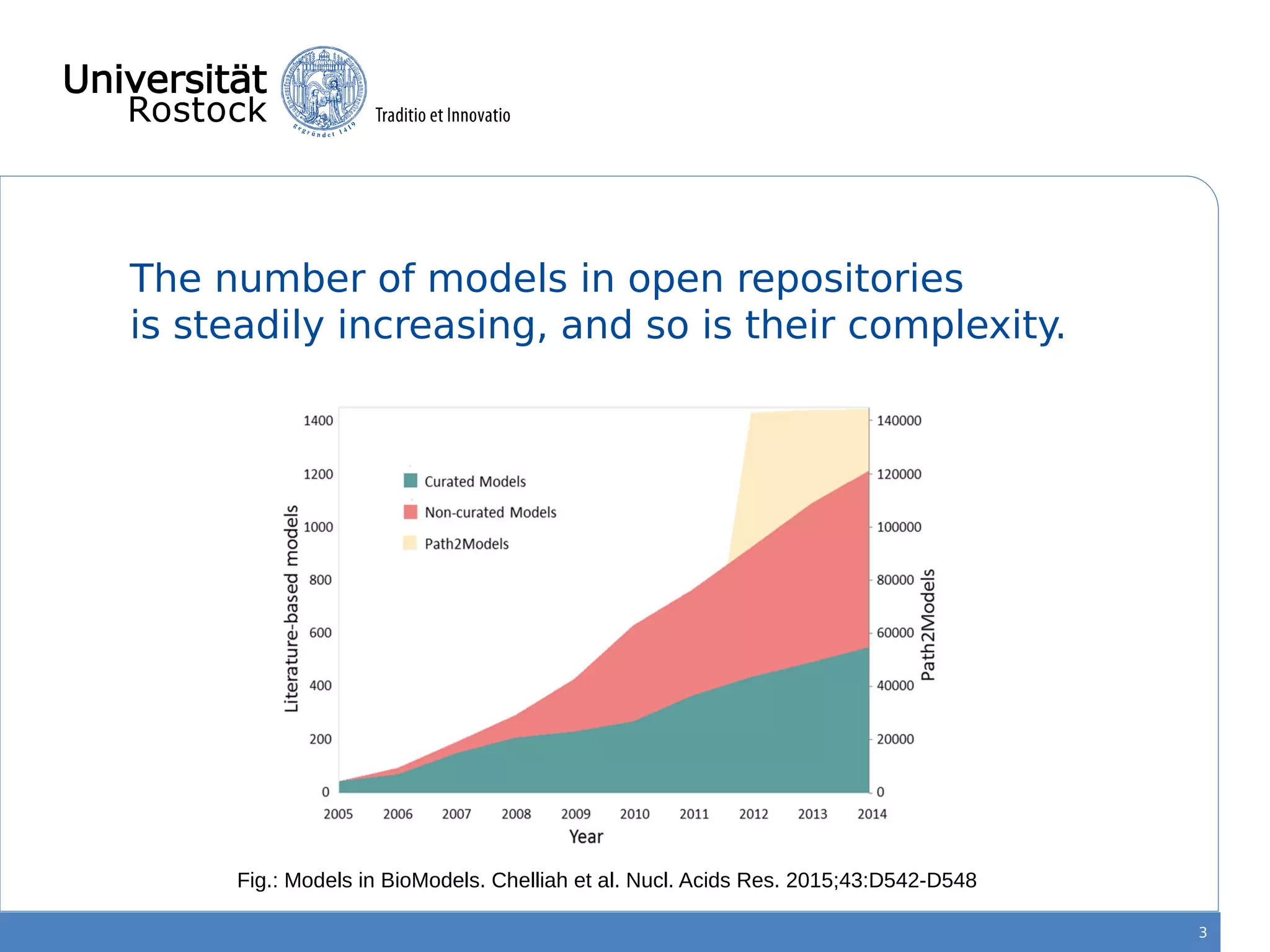
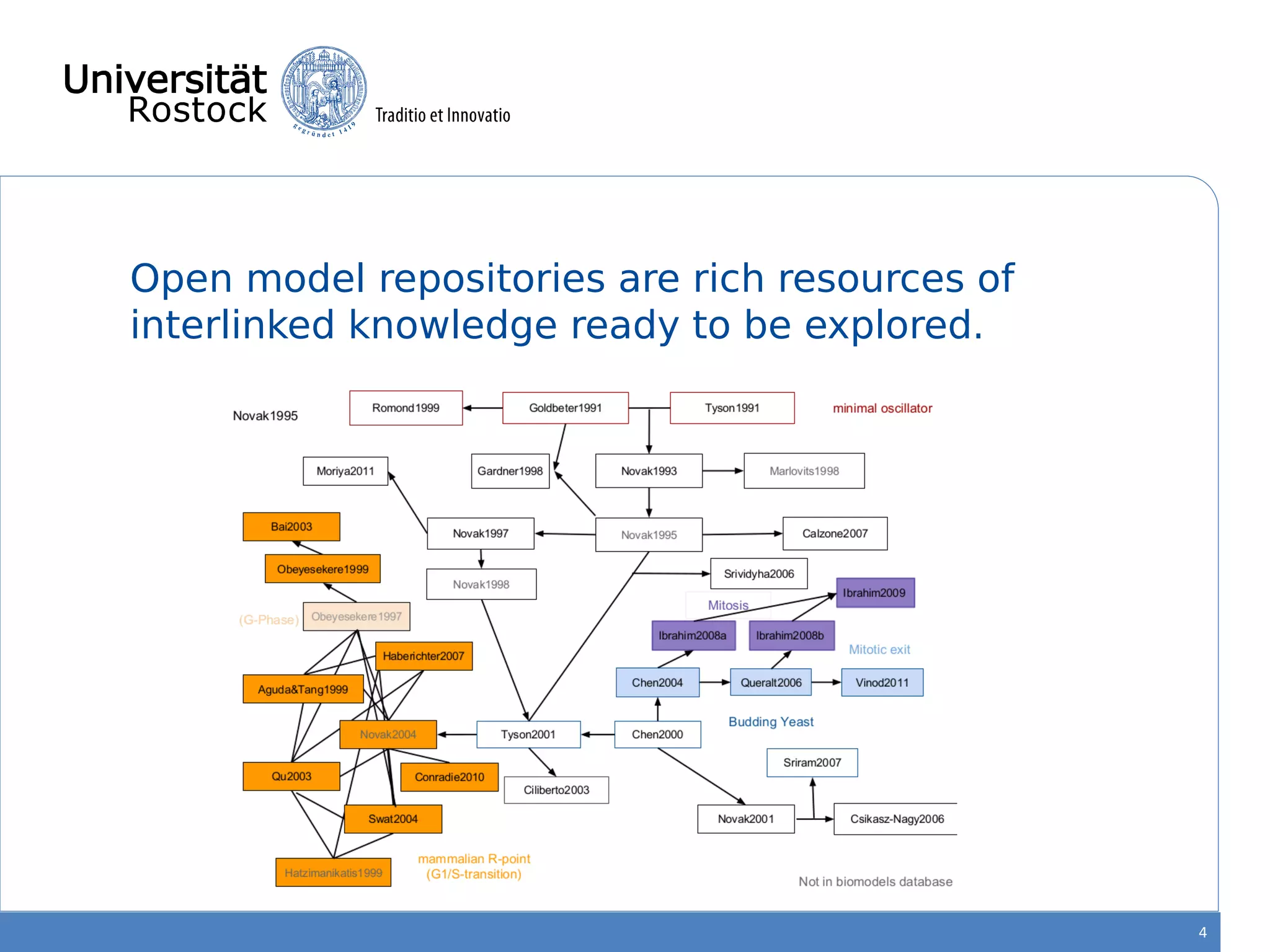
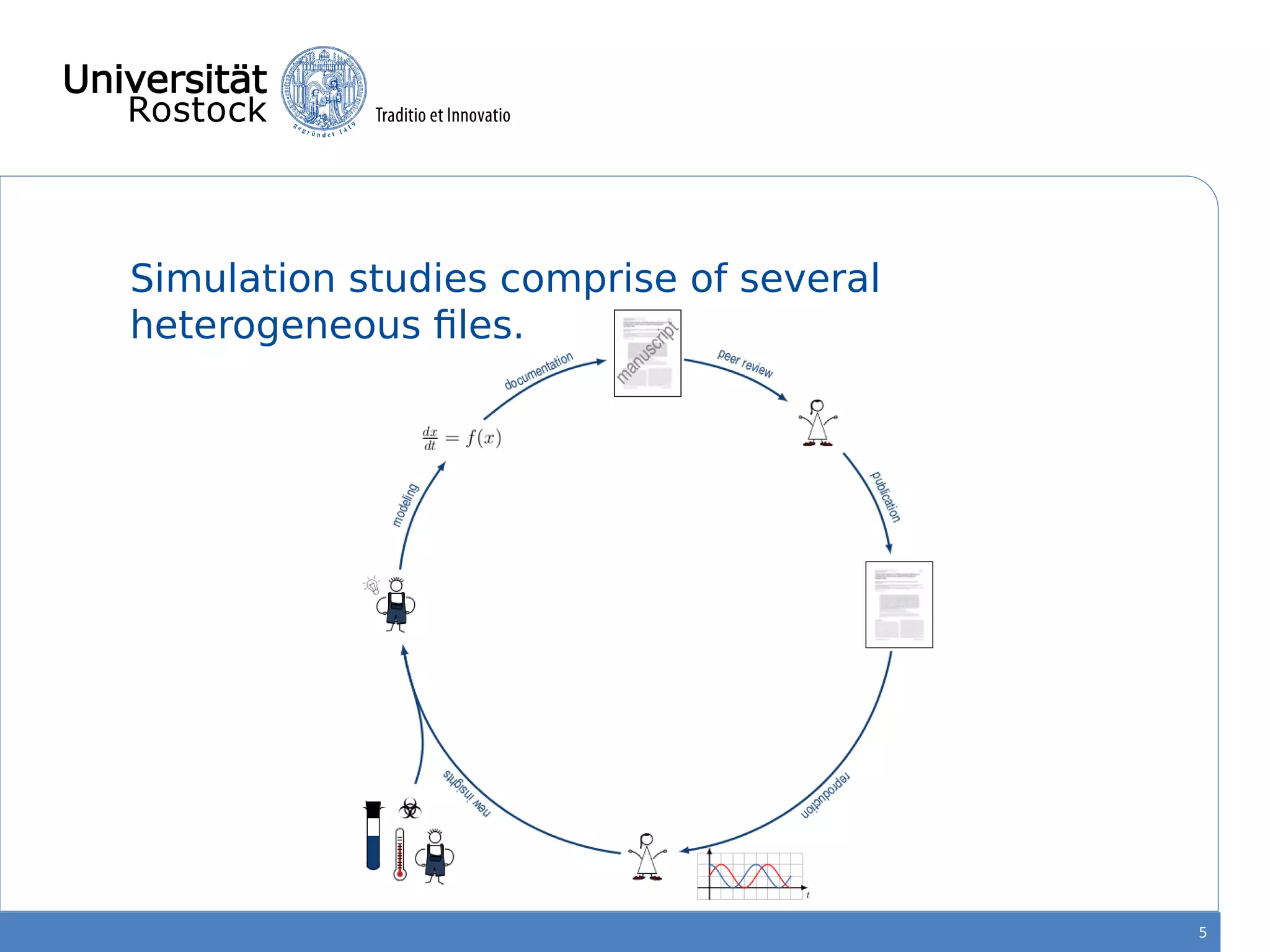
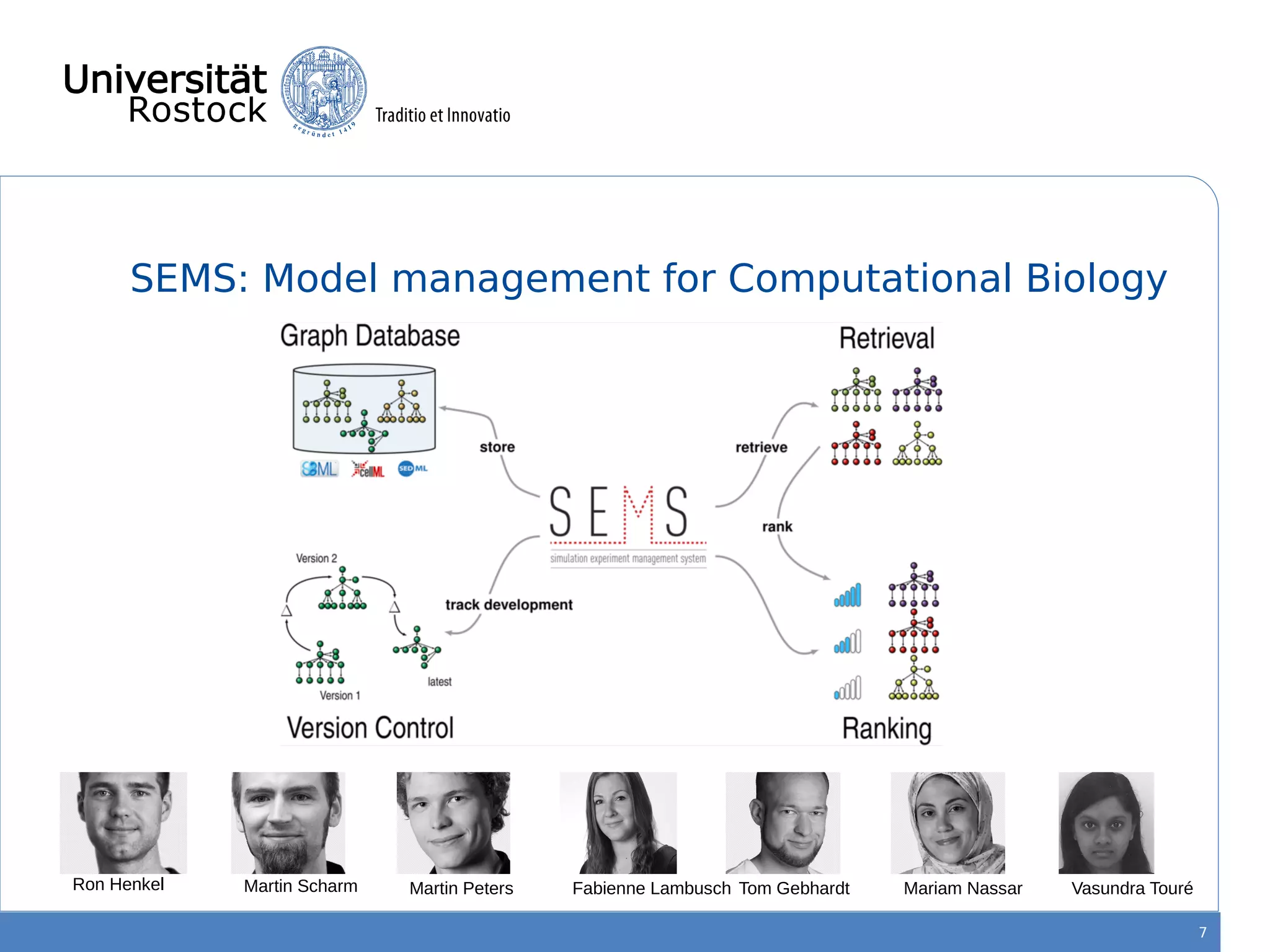
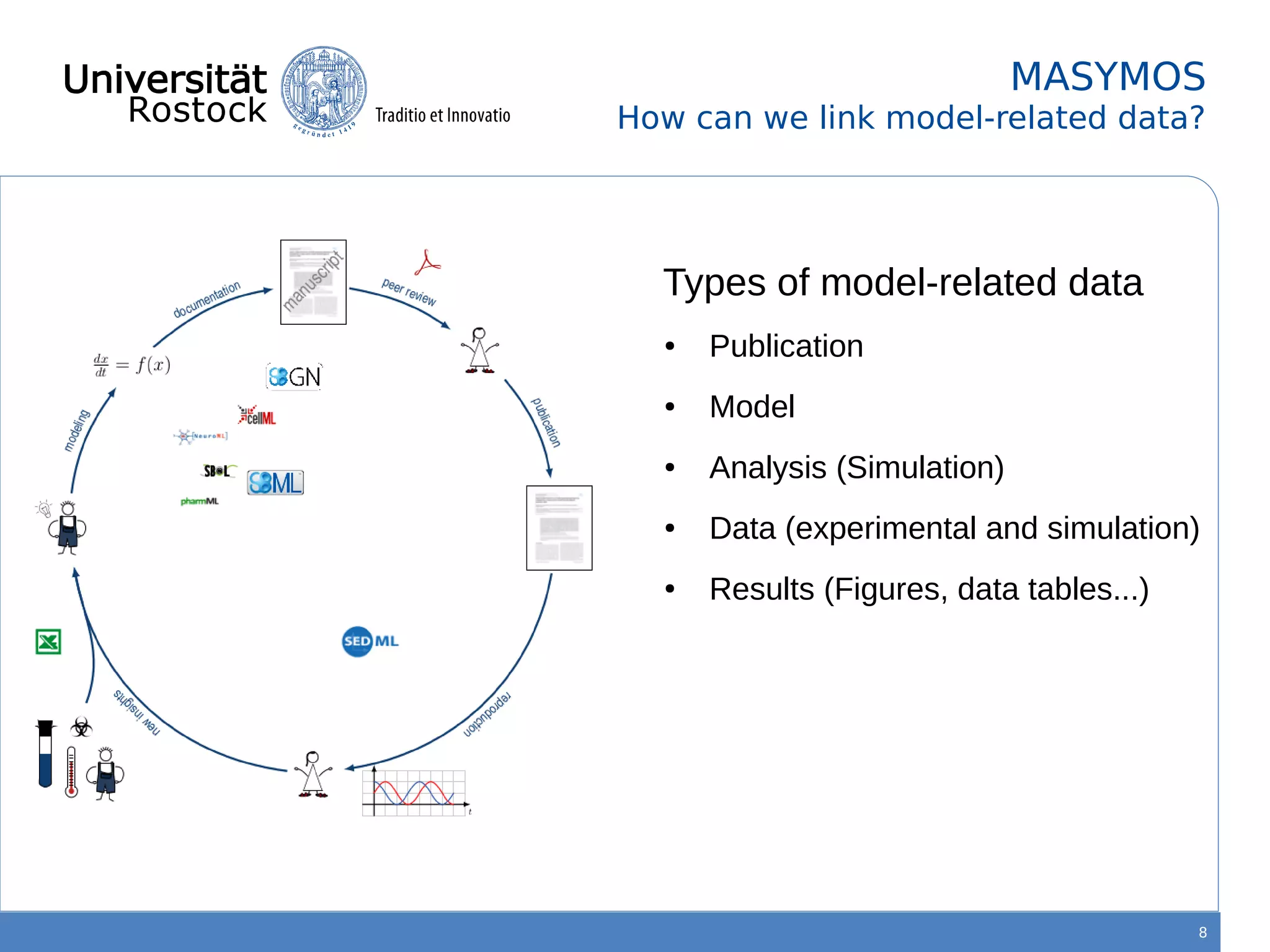
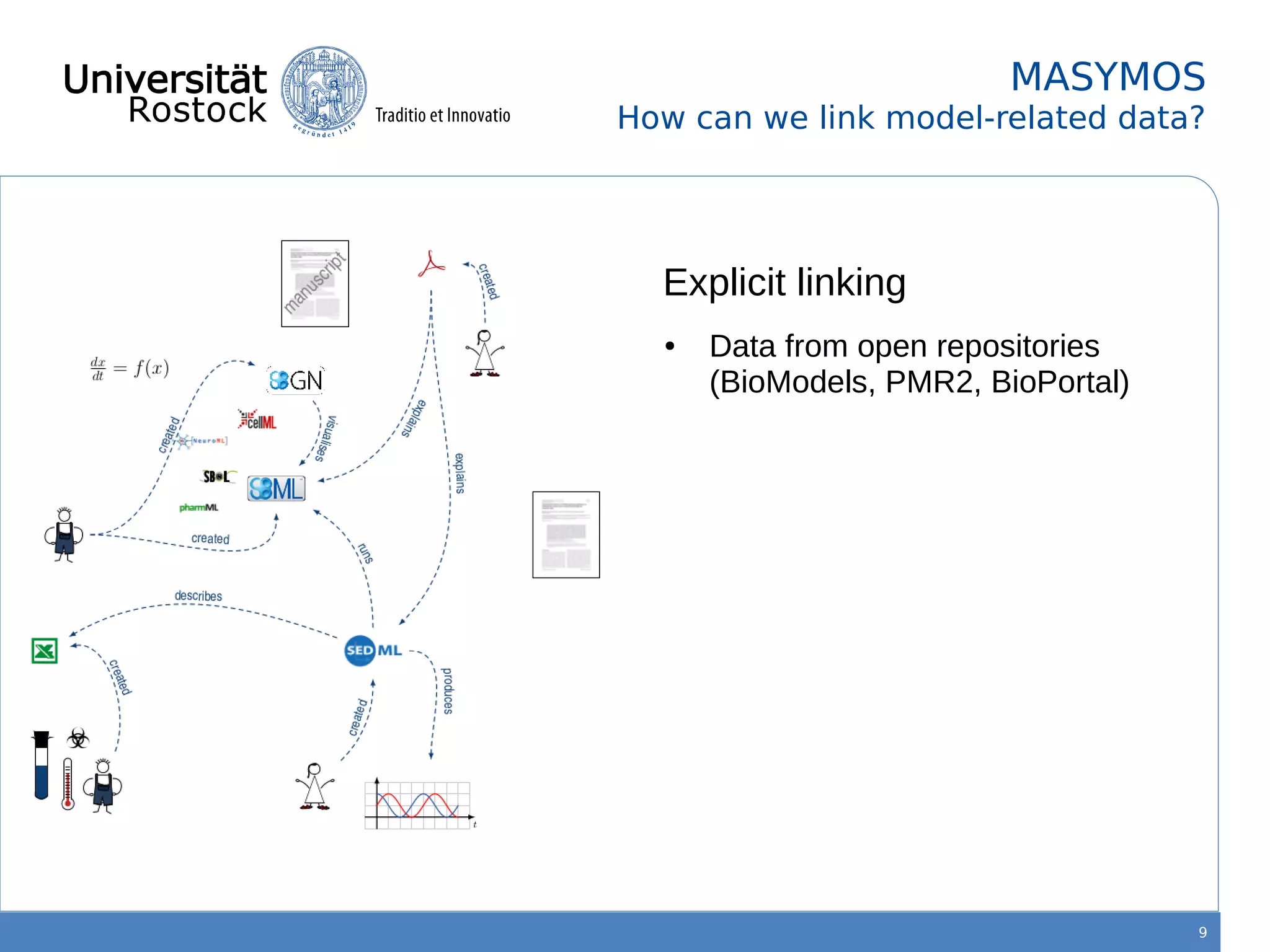
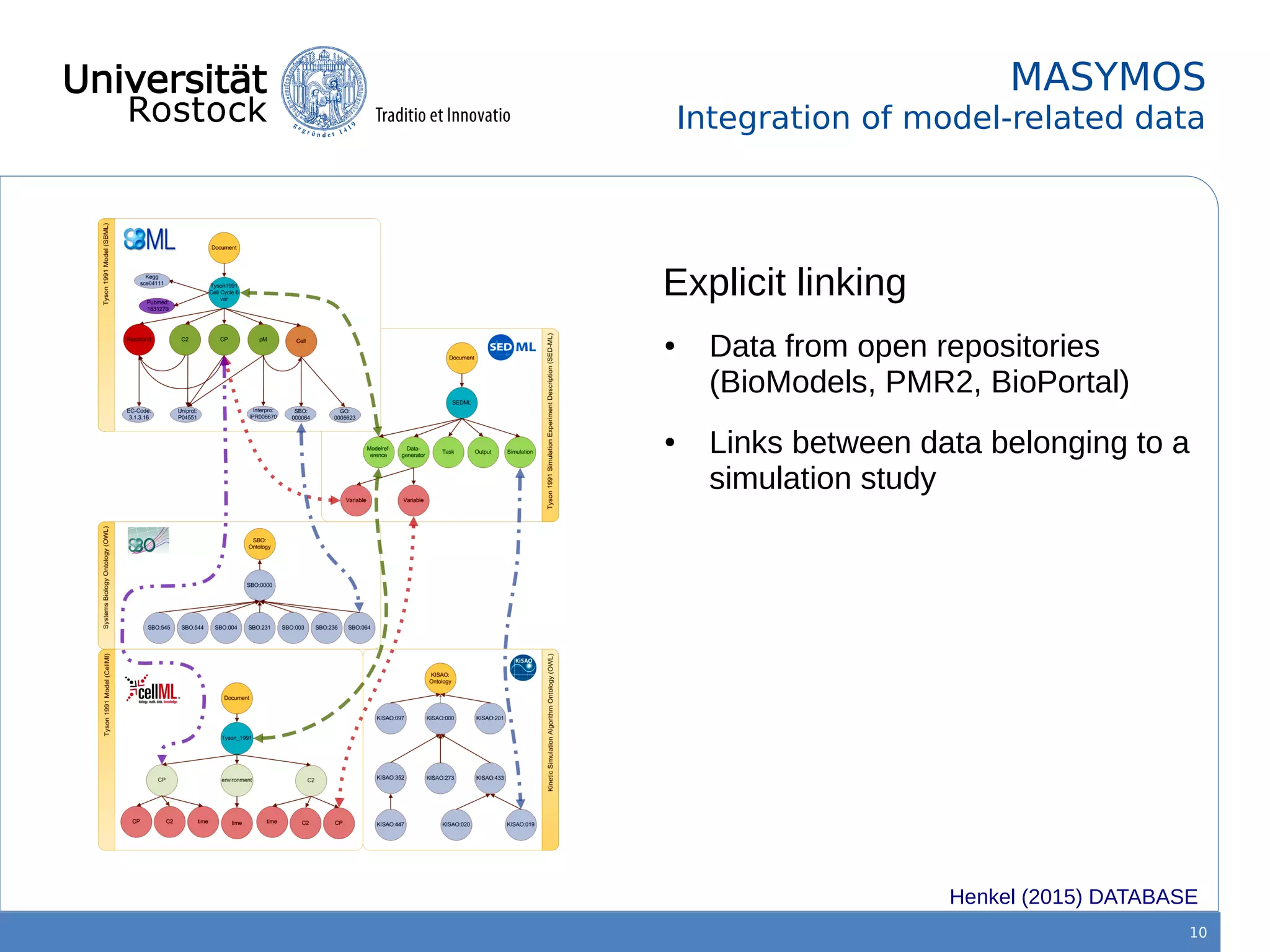
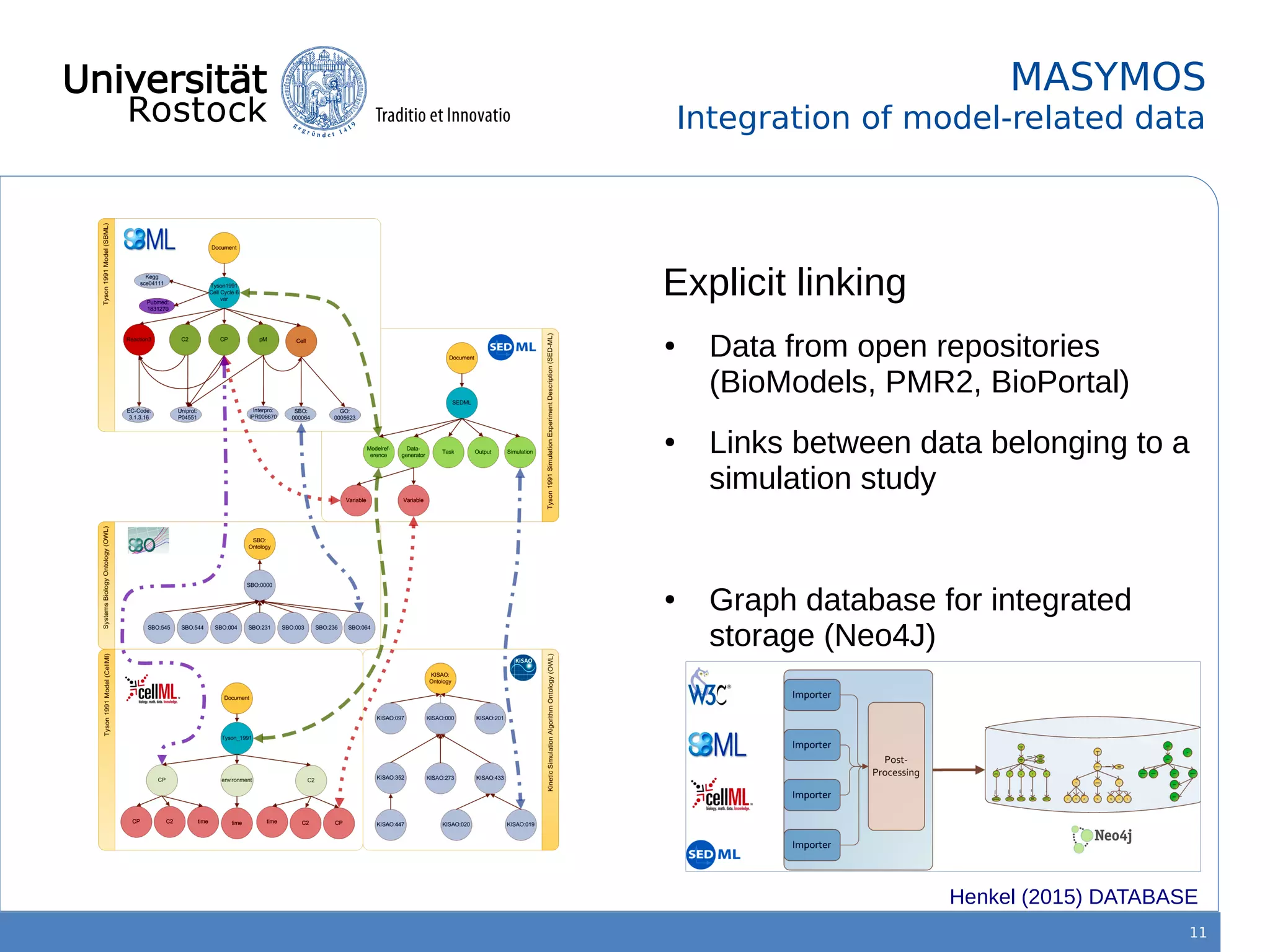
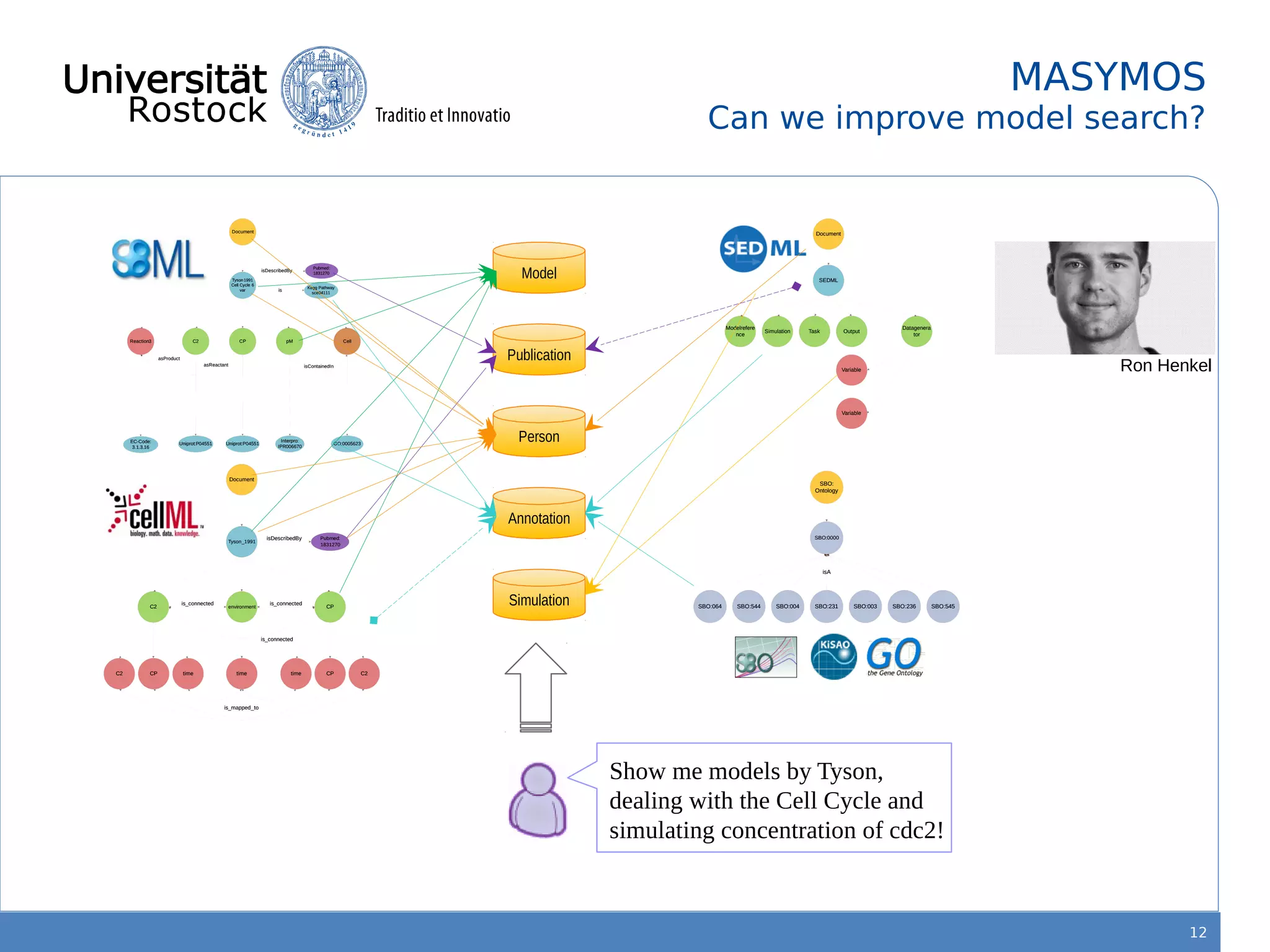
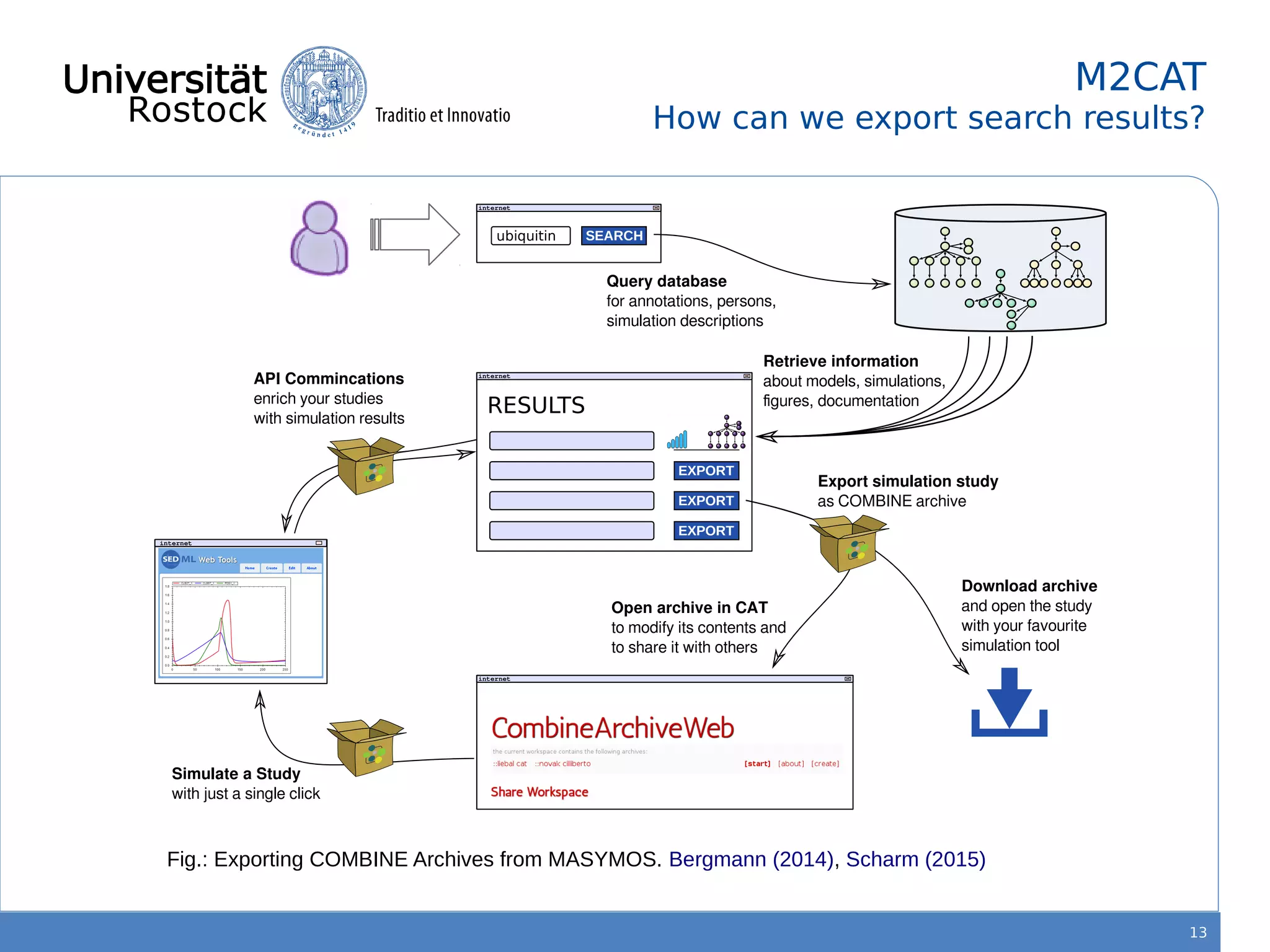
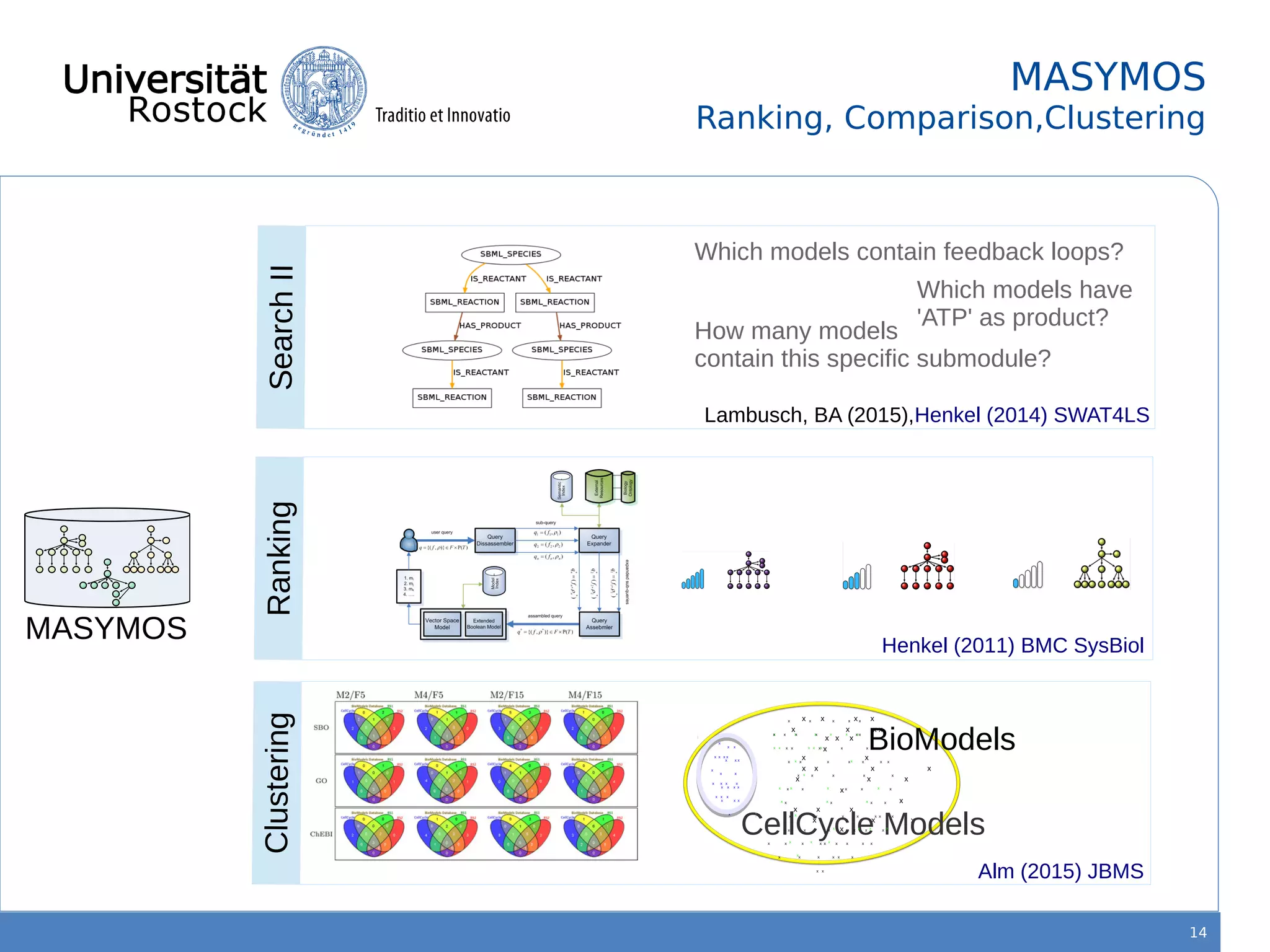
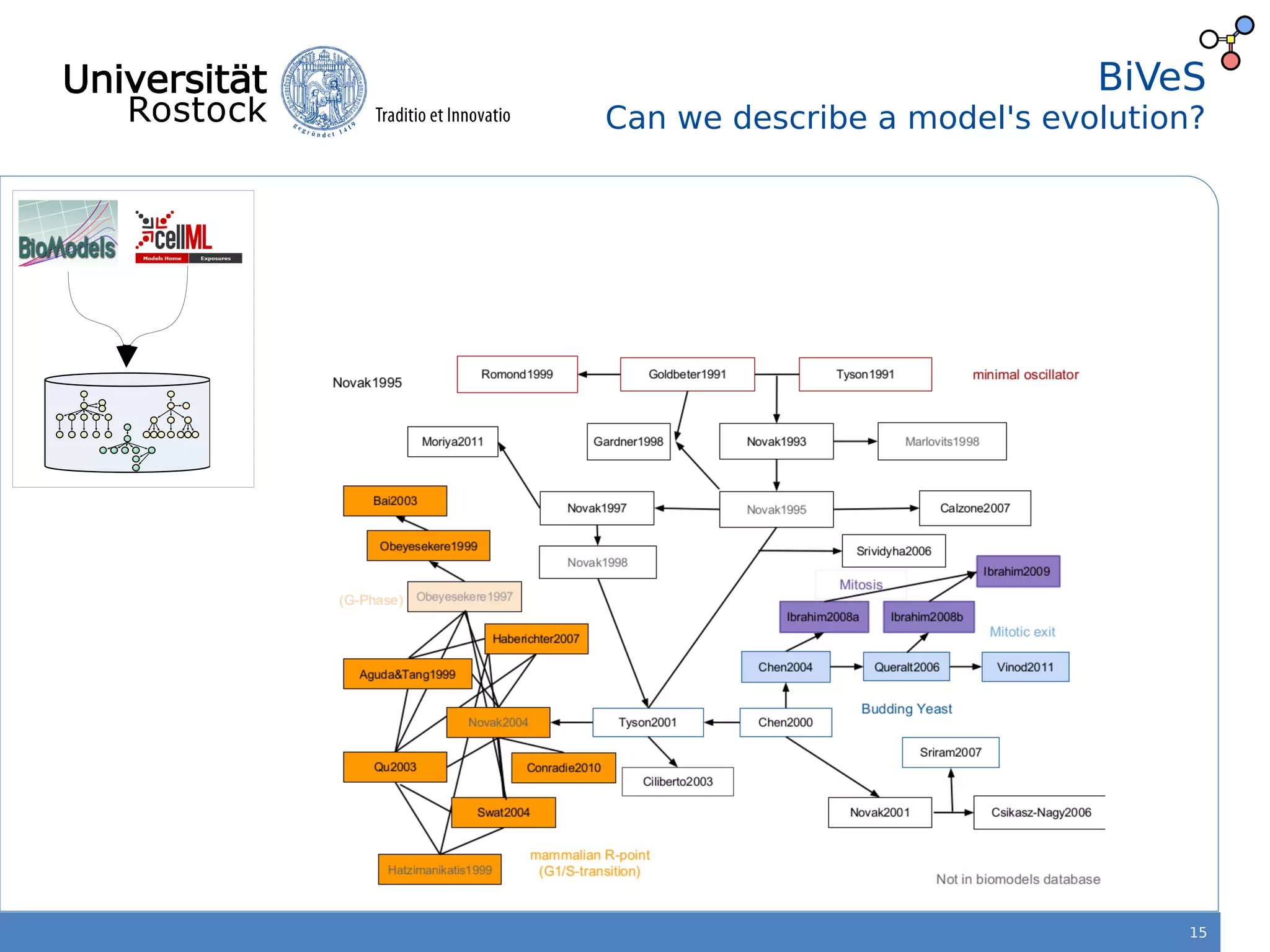
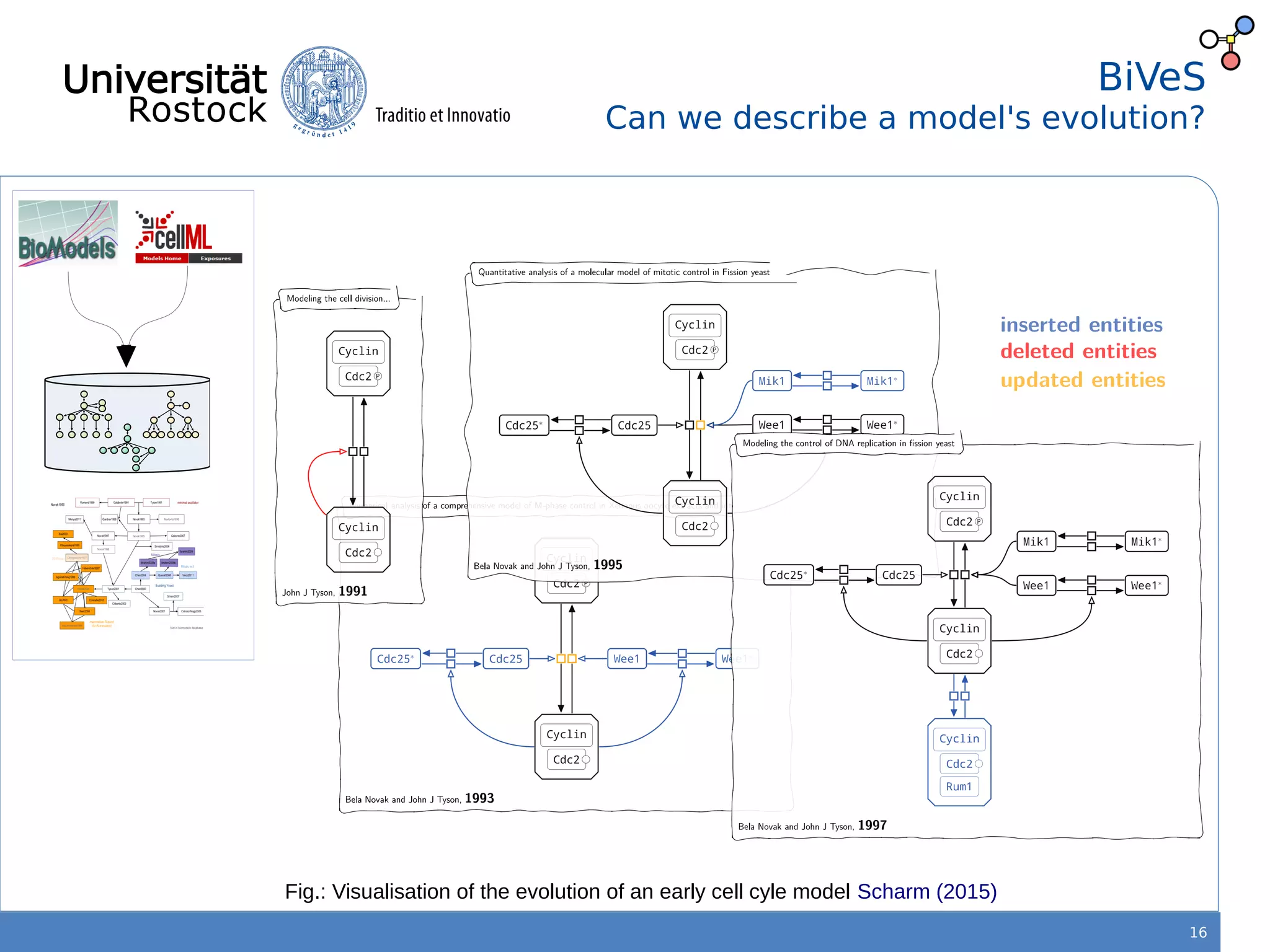
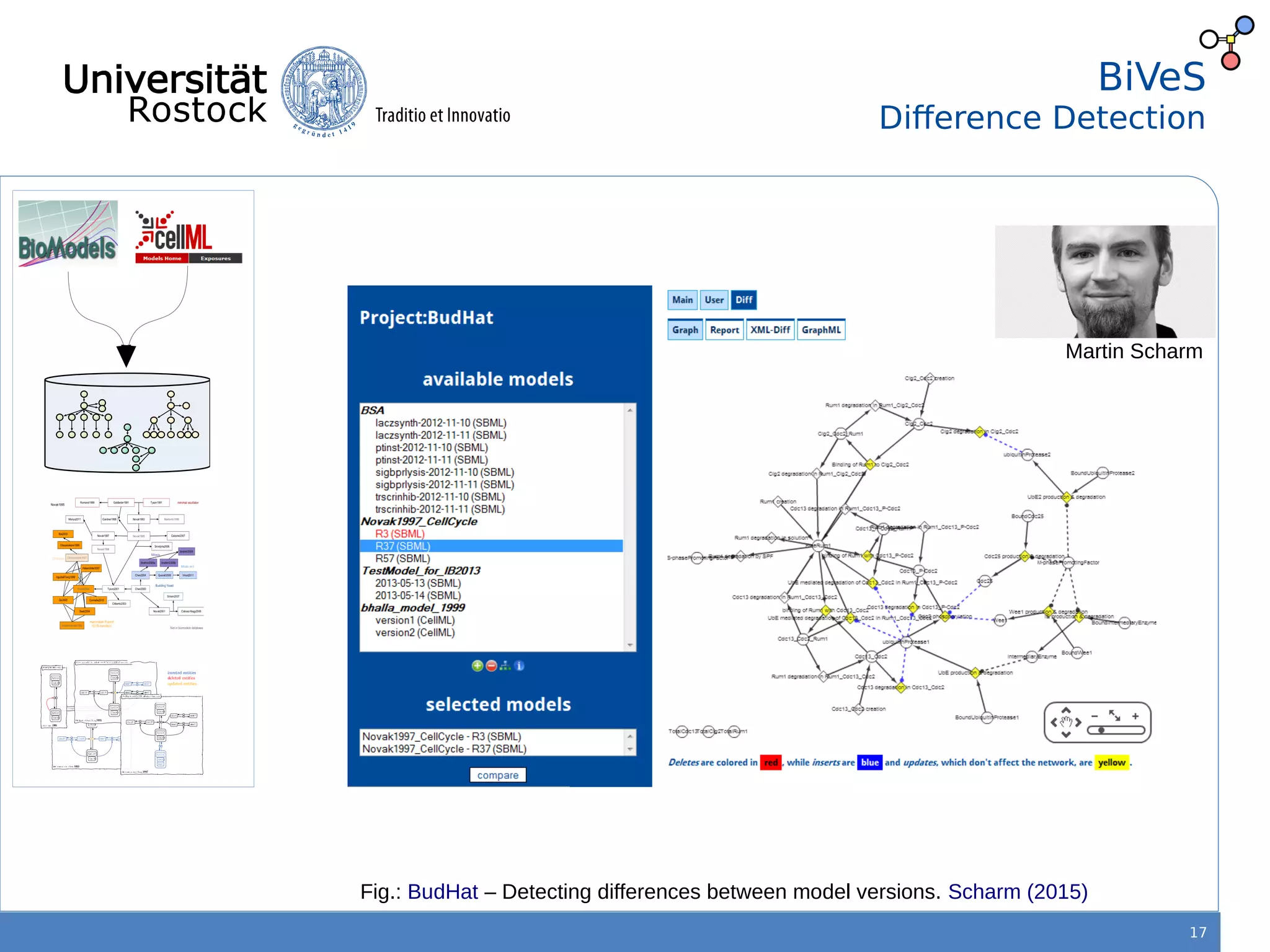
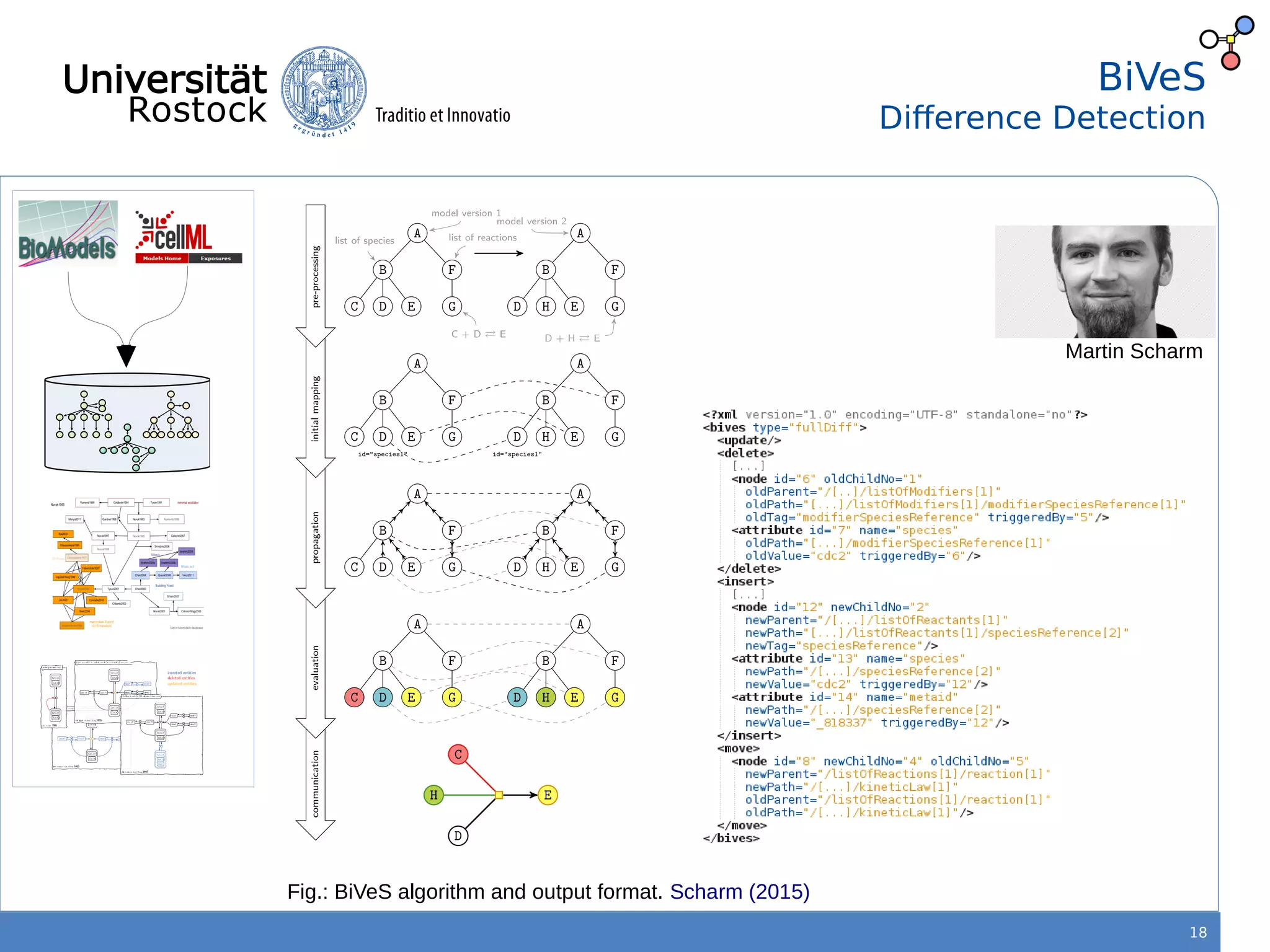
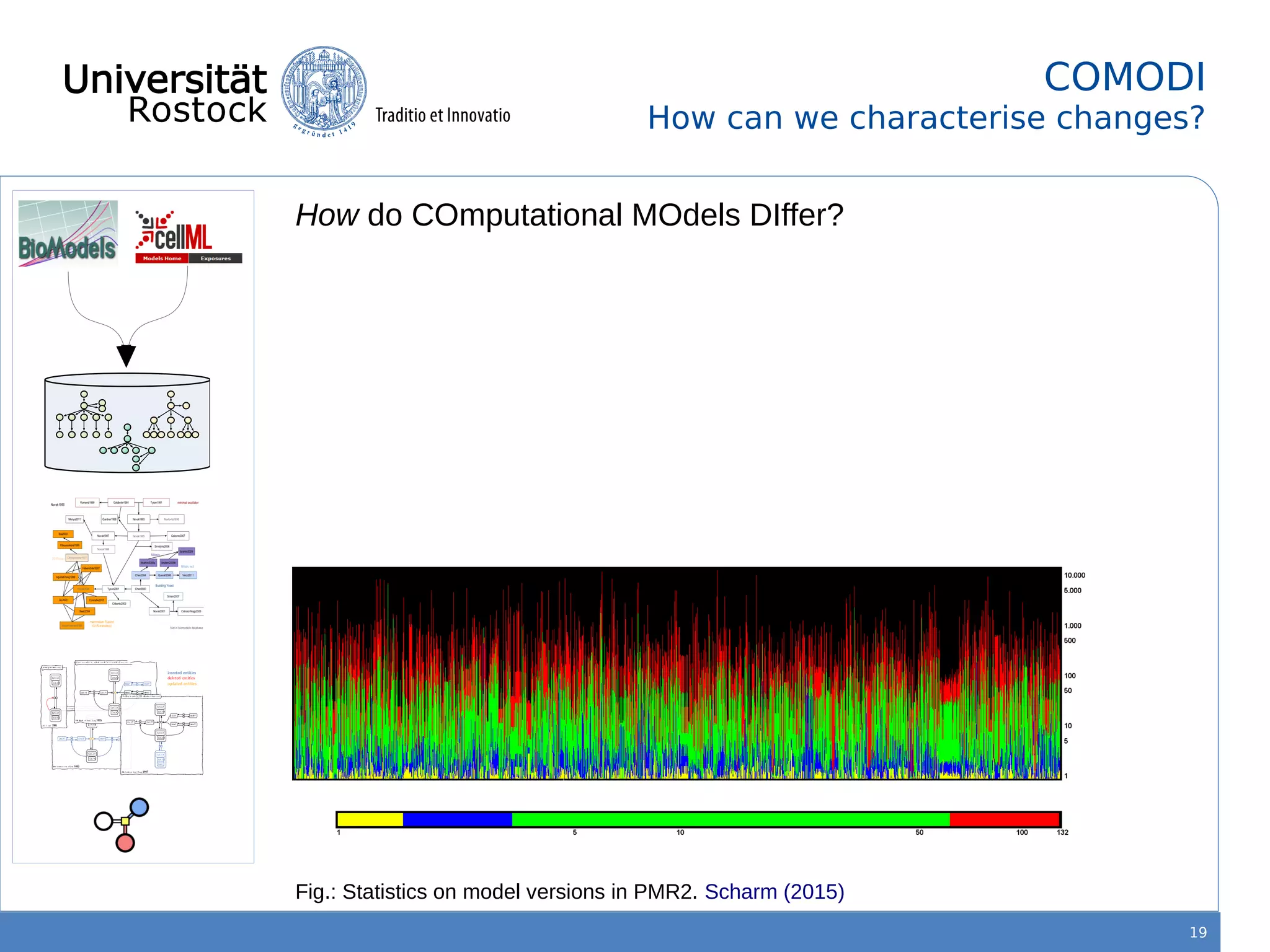
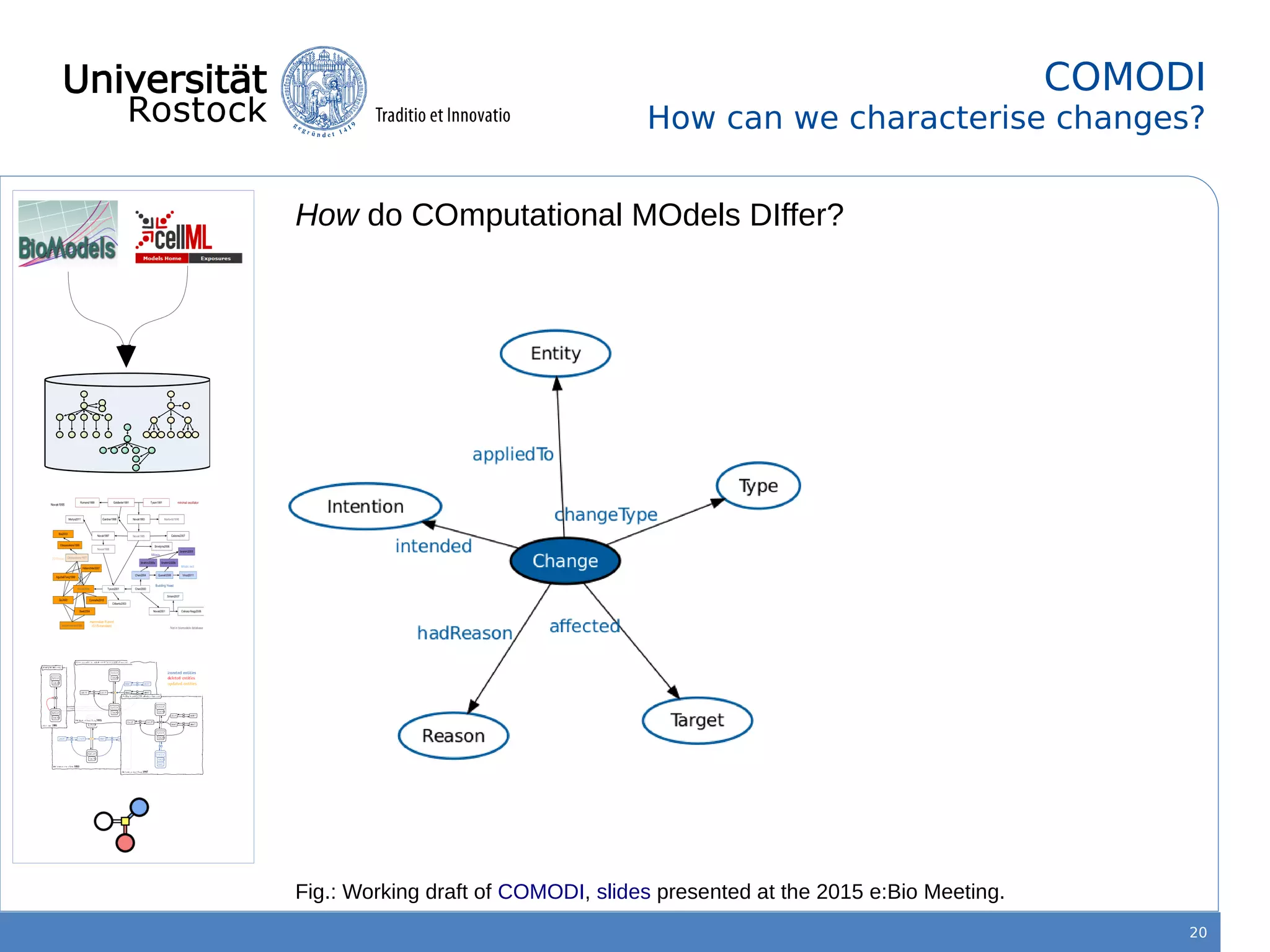
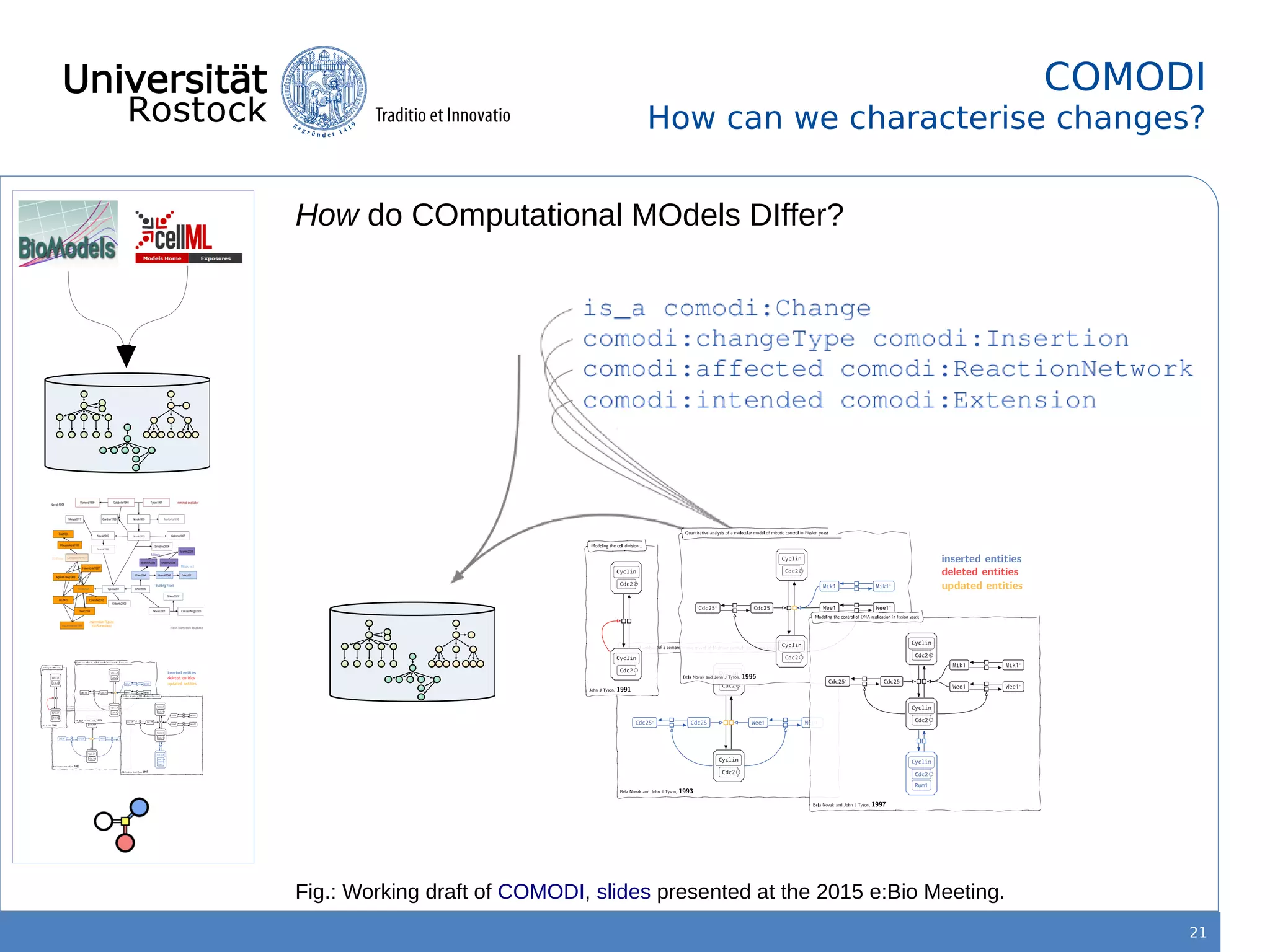
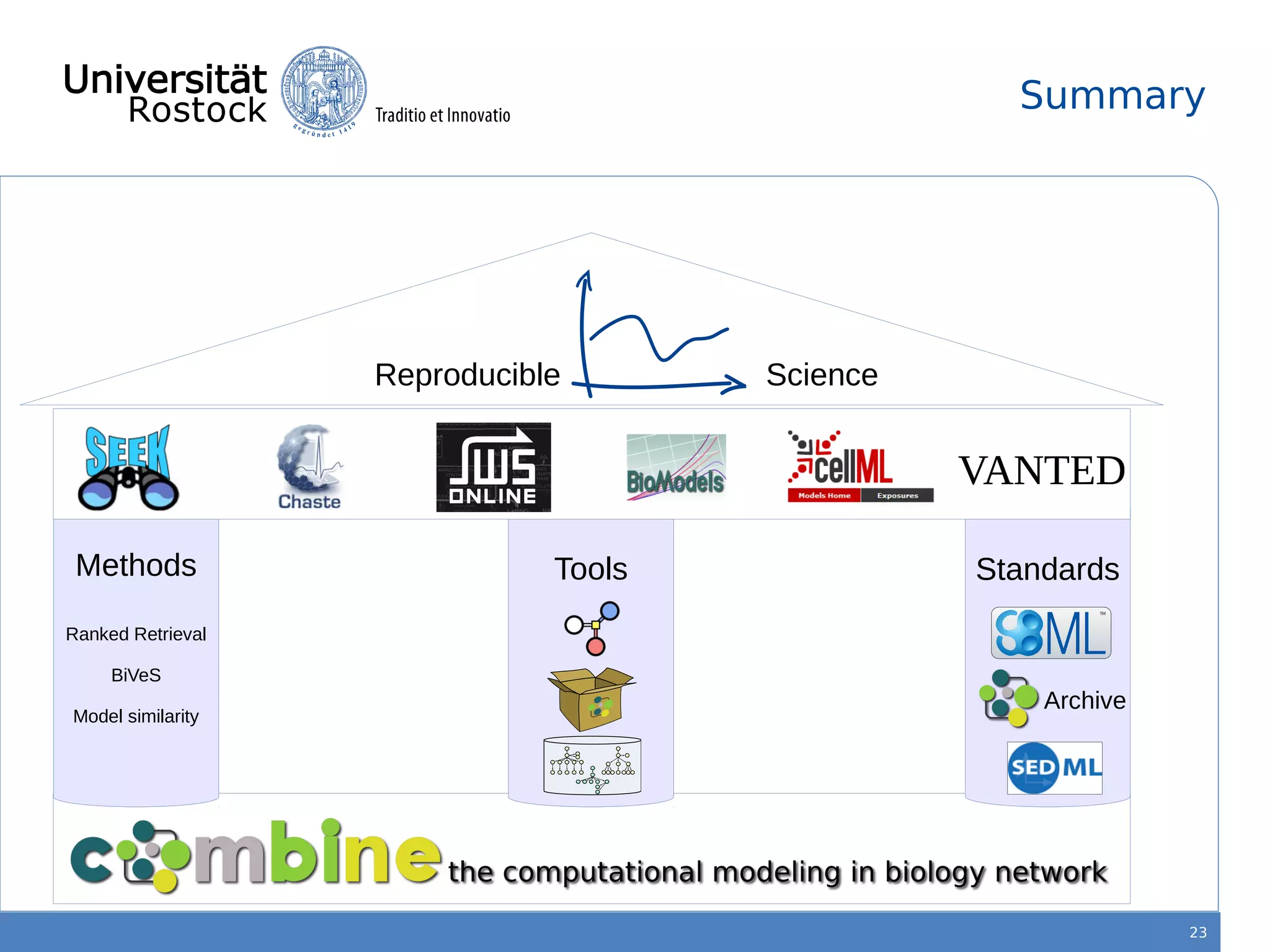
This document discusses improving reproducibility of simulation studies in computational biology through better management of simulation models and data. The SEMS project aims to develop standards and tools to link related data such as publications, models, simulations, results and more. This will be achieved by using graph databases and COMBINE standards to integrate data from various repositories. Tools will be created to search, compare, cluster and visualize models and their evolution over time to enable more reproducible and reusable simulation studies.