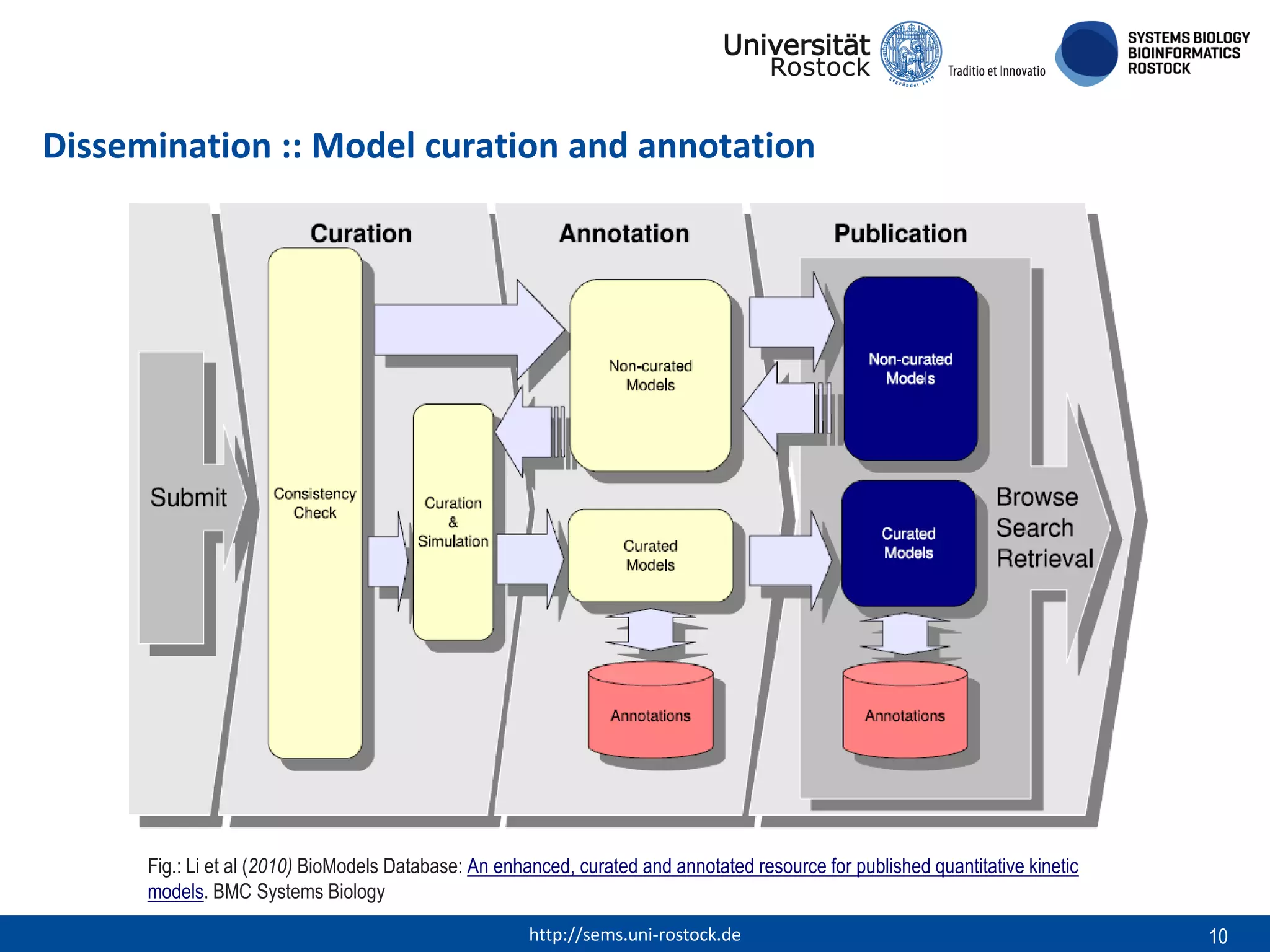
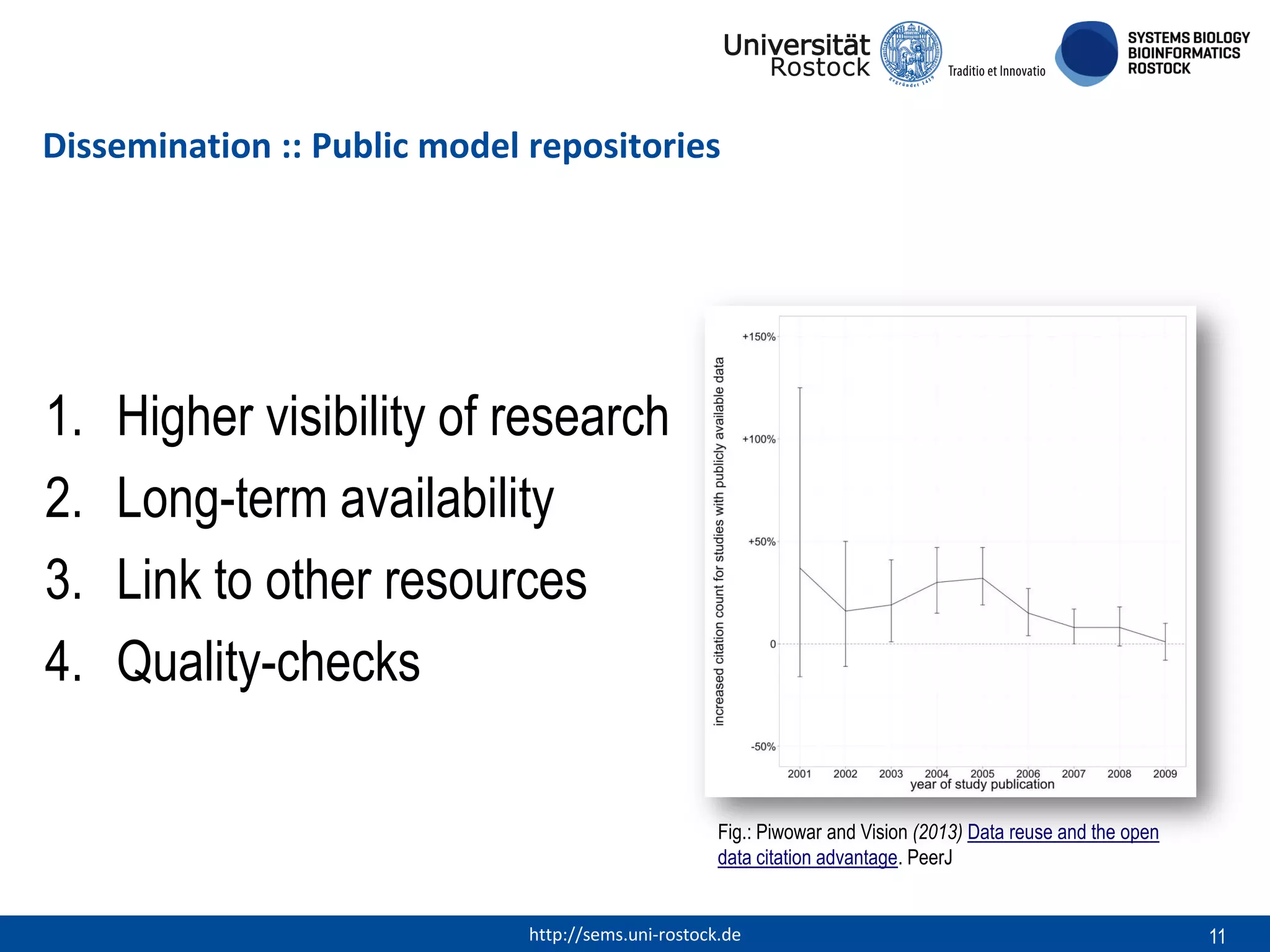
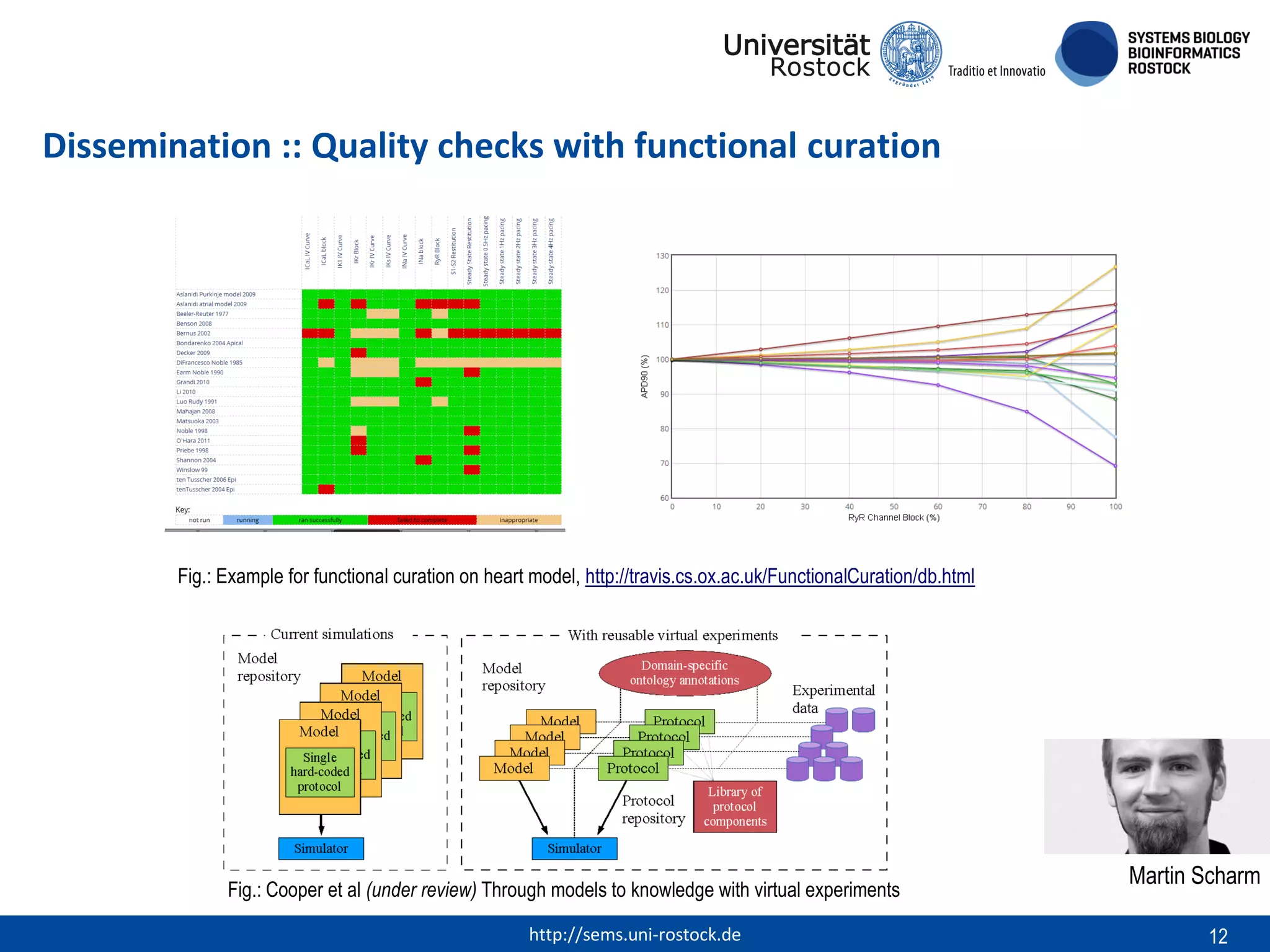
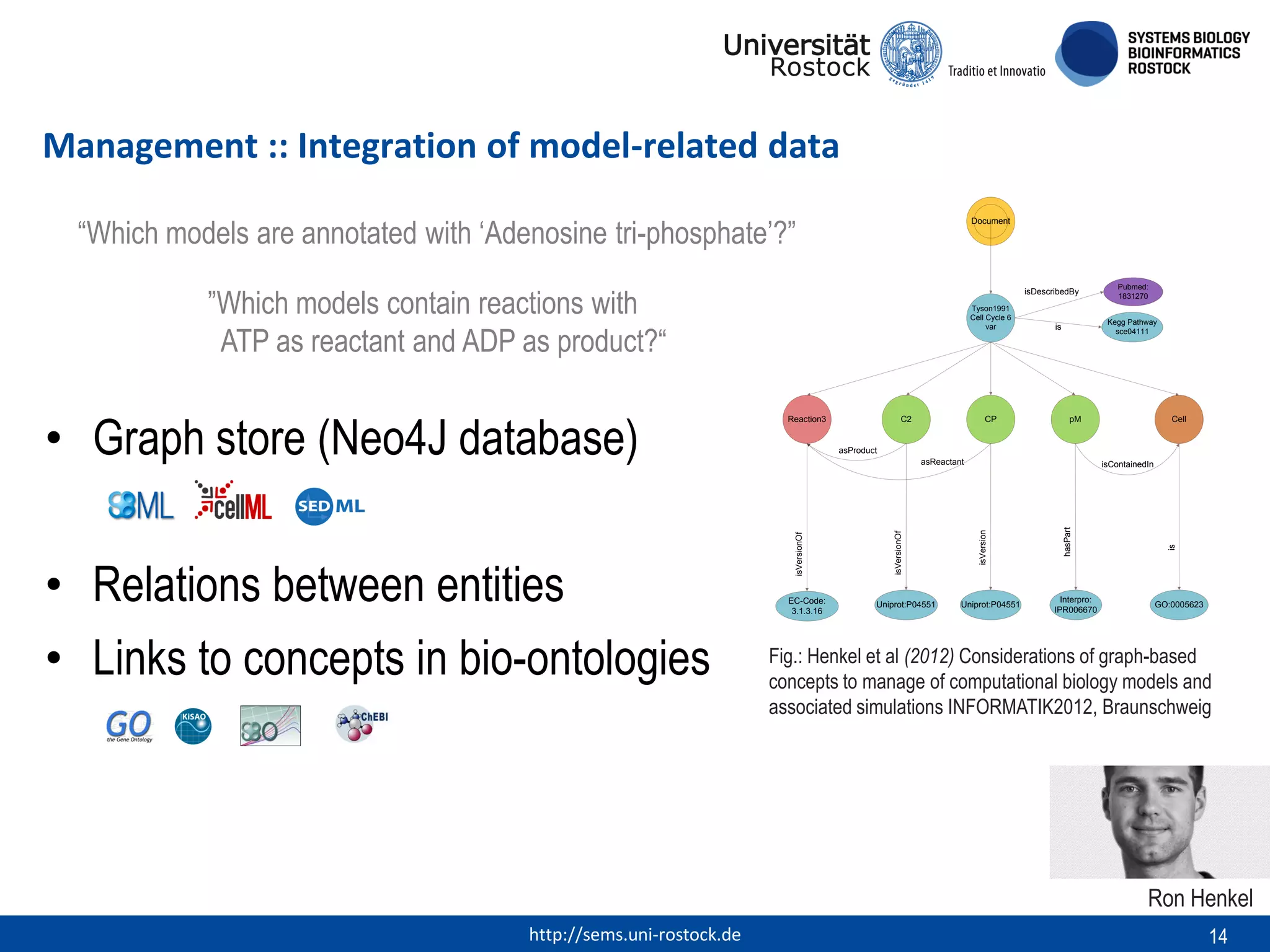
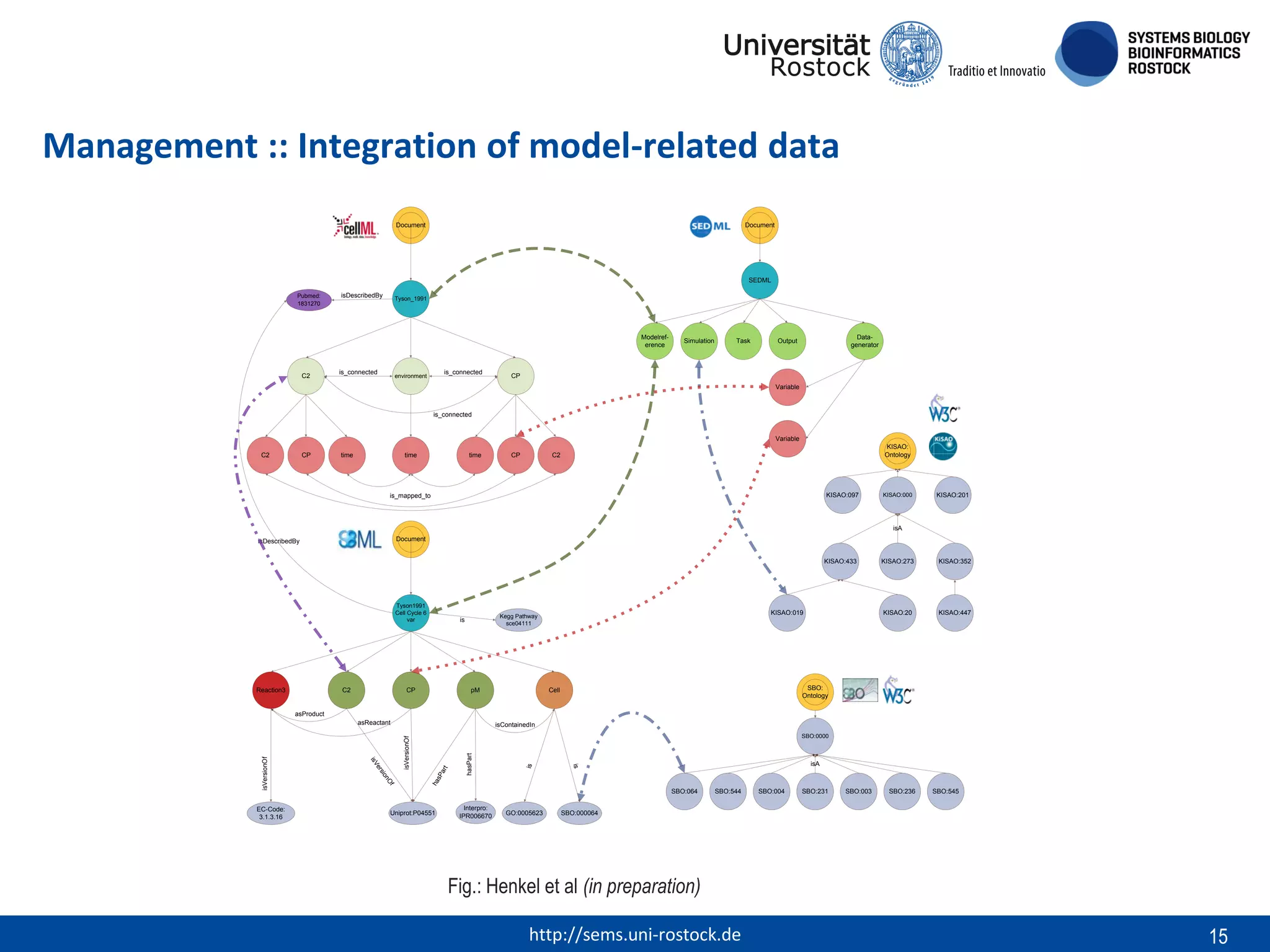
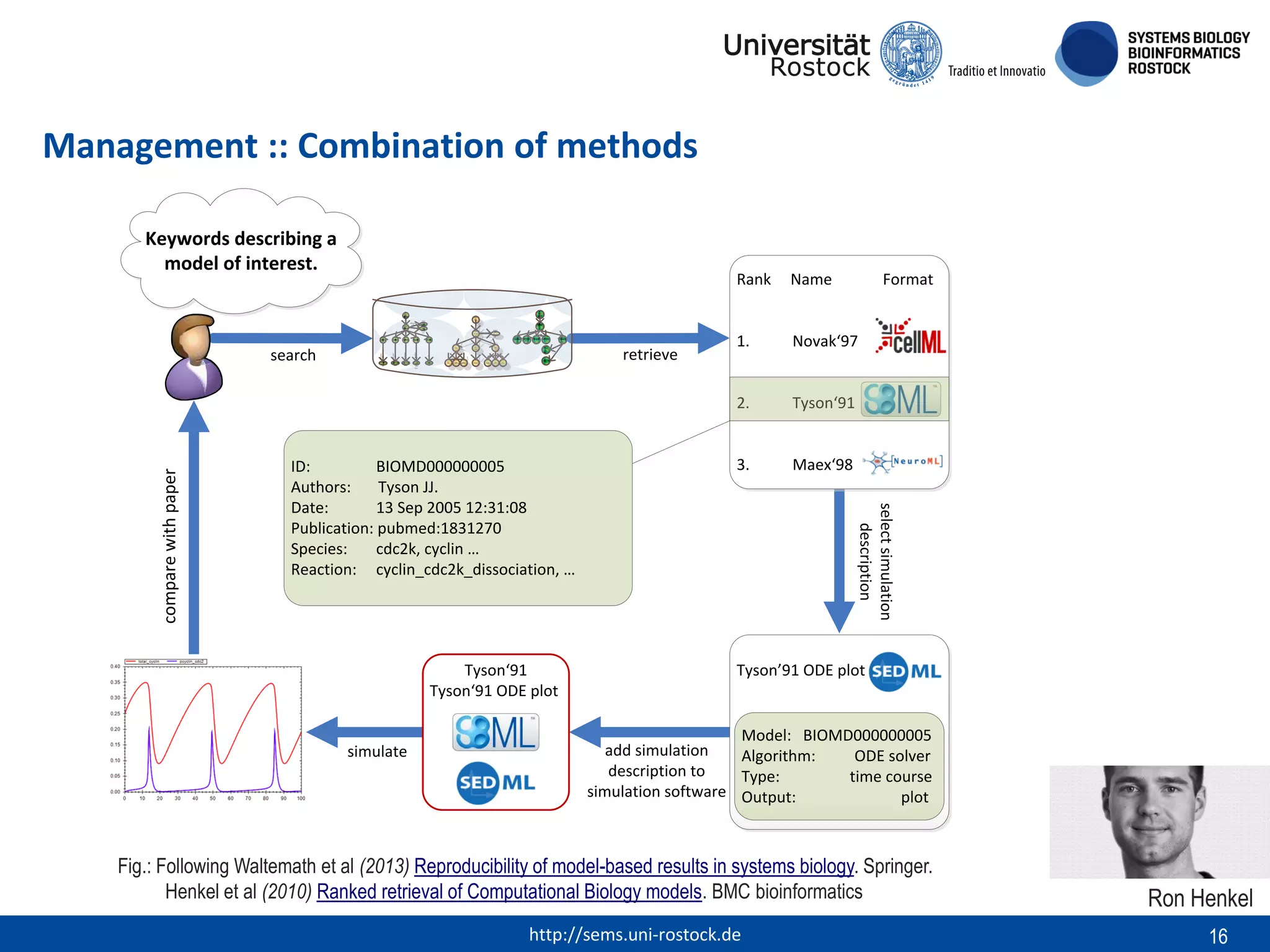
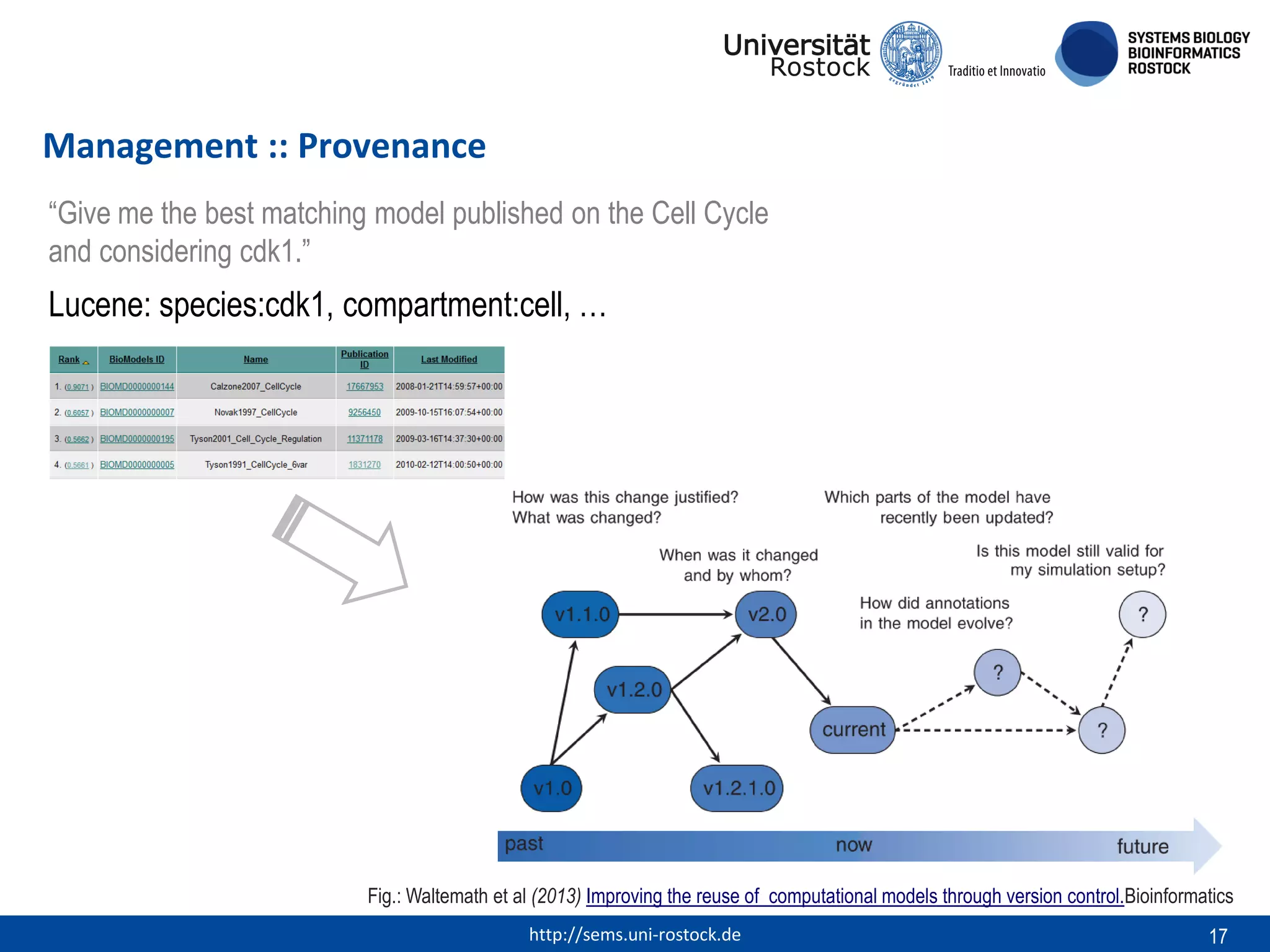
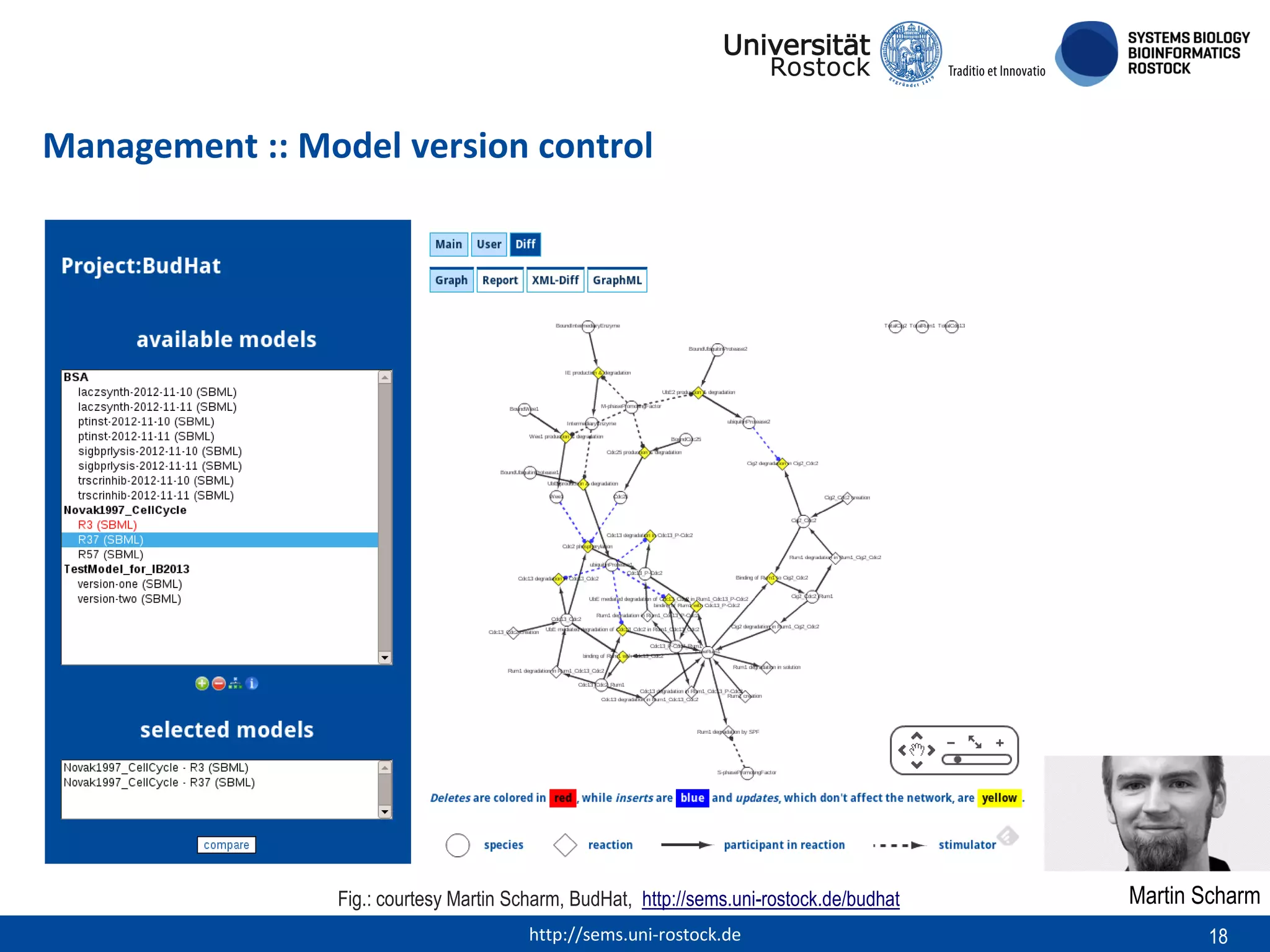
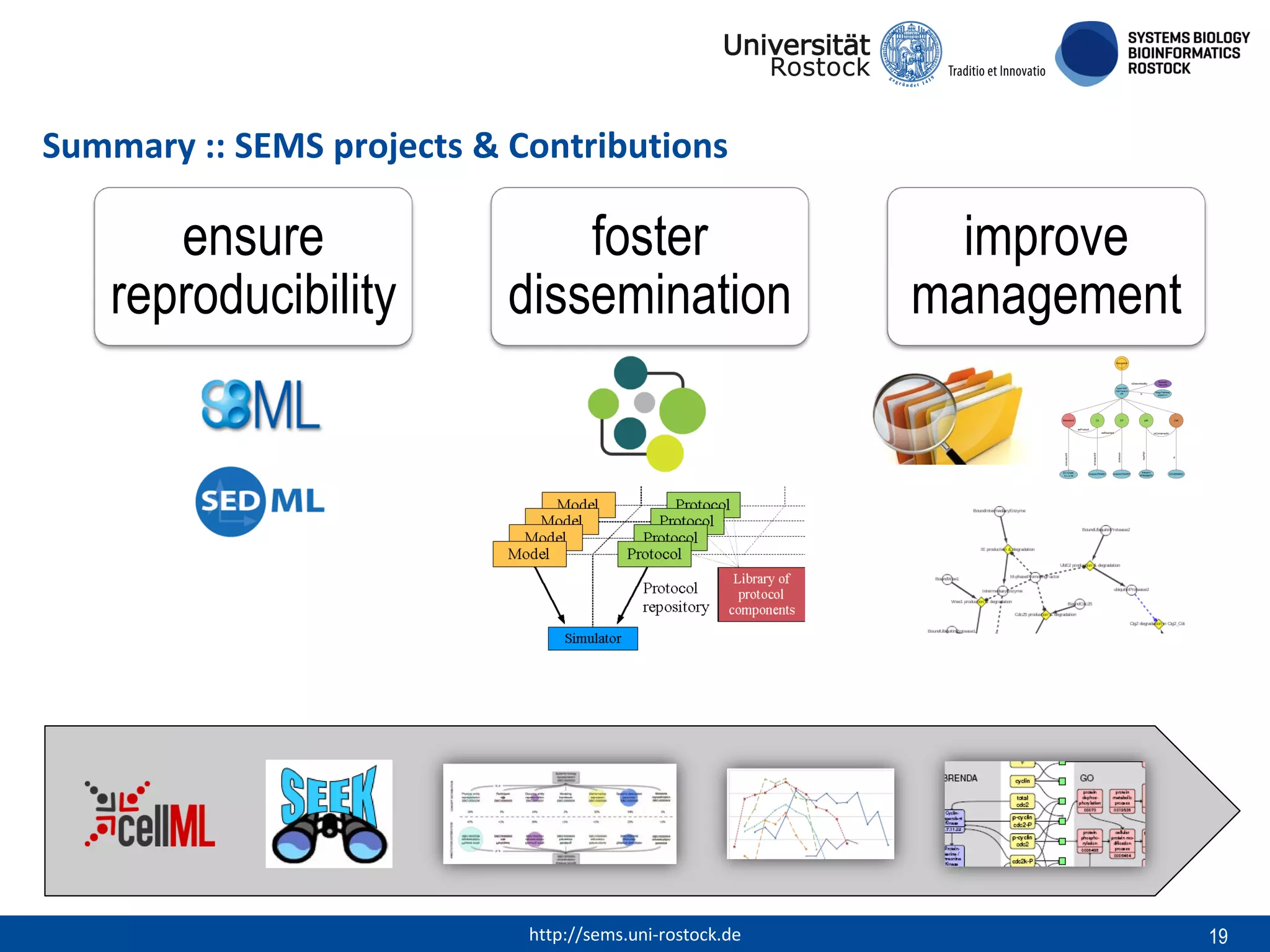
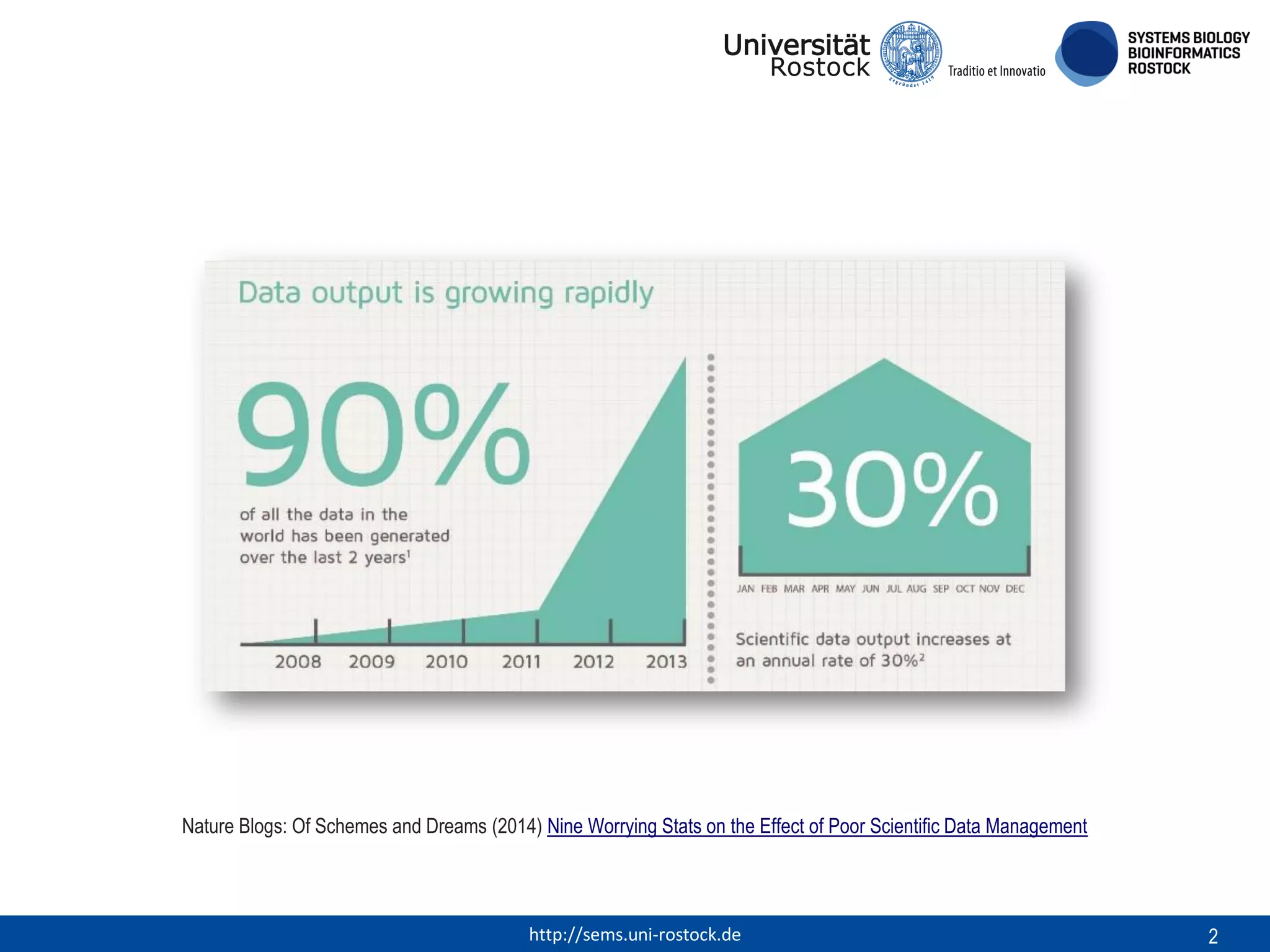
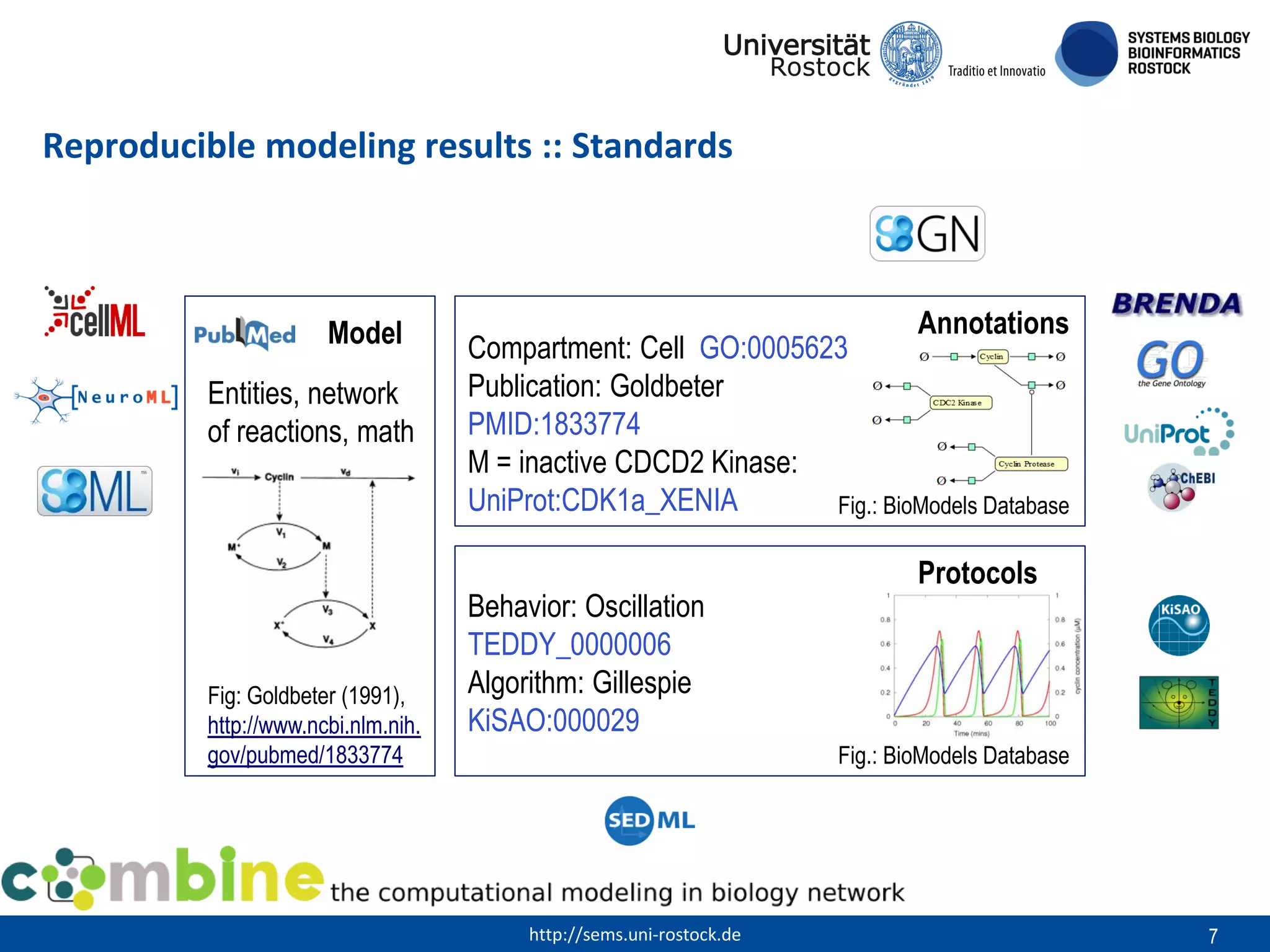
This document discusses challenges in modeling reproducibility, dissemination, and management. It notes that researchers struggle with data management. Standards are needed for reproducible modeling results, including models, annotations, and protocols. Models should be disseminated through public repositories for higher visibility, long-term availability, and quality checks. Management of models and related data can be improved through integration into graph databases linked to ontologies, as well as version control systems. The SEMS projects aim to address these issues to foster dissemination, ensure reproducibility, and improve management of computational models.




![Outline
reproducibility
dissemination
management
[Quantitative] models will be only as useful as their access and reuse
is easy for all scientists.
Nicolas Le Novère (2006) Model storage, exchange and integration. BMC Neuroscience
http://sems.uni-rostock.de
9](https://image.slidesharecdn.com/waltemath2014braunschweig-140303041538-phpapp02/75/Reproducibility-dissemination-and-management-of-modeling-results-9-2048.jpg)