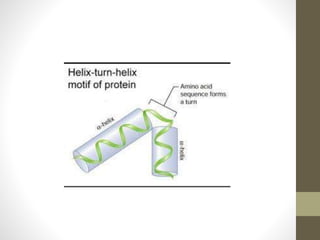
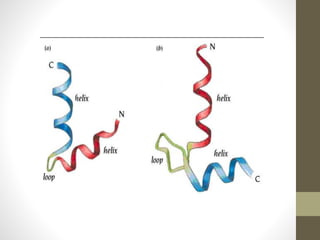
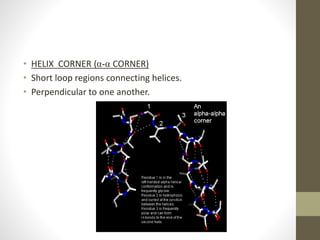
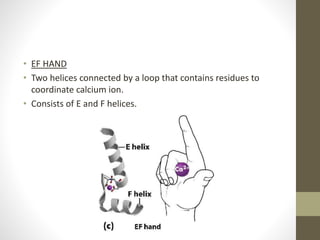
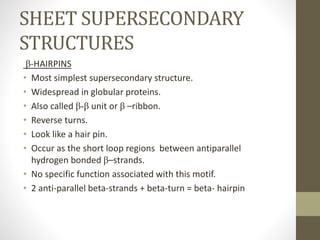
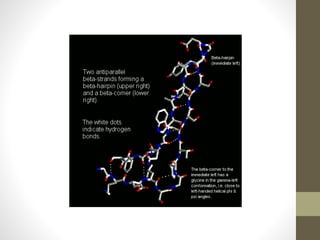
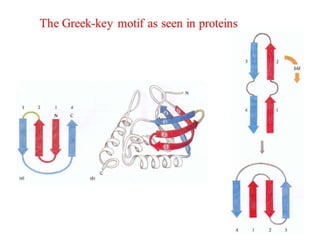
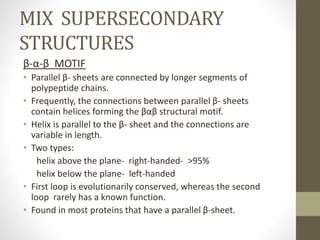
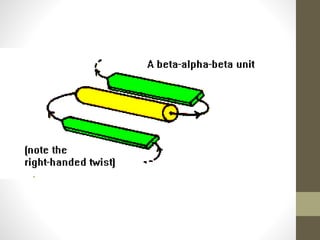
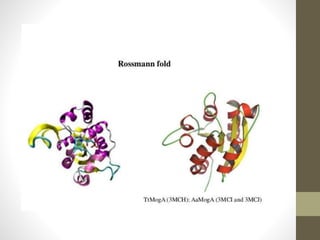
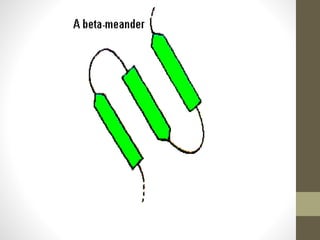
The document discusses various supersecondary structures of proteins, which are intermediate structures between secondary and tertiary protein structures. It describes several common motifs composed of two or more secondary structures, such as helix-turn-helix, helix-loop-helix, beta-hairpins, and the Rossmann fold. These motifs are building blocks that occur frequently in protein structures and are associated with specific functions like DNA binding. The document provides detailed examples and diagrams of different supersecondary structure motifs involving helices, strands, and their combinations.